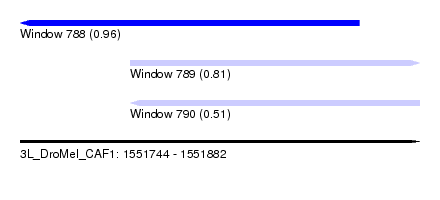
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,551,744 – 1,551,882 |
| Length | 138 |
| Max. P | 0.957753 |

| Location | 1,551,744 – 1,551,861 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.42 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -26.68 |
| Energy contribution | -27.30 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
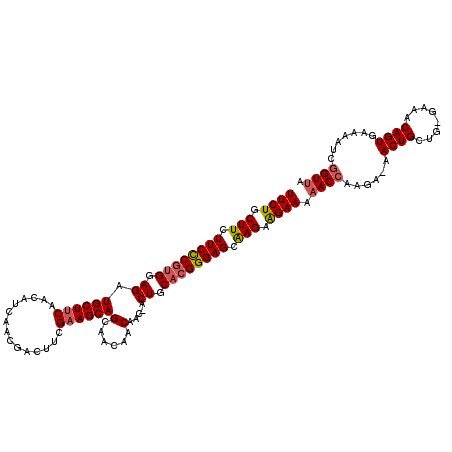
>3L_DroMel_CAF1 1551744 117 - 23771897 UUCUGCUUCUUCCCGUGGAGAUGCUUCAACAUCAACGACUUCGAAGCAGCAACAACAAC-ACUGCACGGGAGCGAGAAGAAAAACCAAGA-AACUGCUG-GAAACAGUGAAAAUCGGUUA ((((.(((.((((((((.((.((((((...............))))))(......)...-.)).)))))))).))).)))).((((....-....((((-....))))(.....))))). ( -35.76) >DroSec_CAF1 9452 117 - 1 UUCUGCUUCUUCCCGUGGAGAUGCUUCAACAUCAACGACUUCGAAGCAGCAACAACAAC-ACUGCACGGGAGCAAGAAGAAAAACCAAGA-AACUGCUG-GAAACAGUGAAAAUCGGUUA ((((.(((.((((((((.((.((((((...............))))))(......)...-.)).)))))))).))).)))).((((....-....((((-....))))(.....))))). ( -35.36) >DroSim_CAF1 9571 117 - 1 UUCUGCUUCUUCCCGUGGAGAUGCUUCAACAUCAACGACUUCGAAGCAGCAACAACAAC-ACUGCACGGGAGCAAGAAGAAAAACCAAGA-AACUGCUG-GAAACAGUGAAAAUCGGUUA ((((.(((.((((((((.((.((((((...............))))))(......)...-.)).)))))))).))).)))).((((....-....((((-....))))(.....))))). ( -35.36) >DroYak_CAF1 10284 117 - 1 UUCUGCUUCUUCUCGCGGAGAUGCUGCAACA---ACAACUGCGAAGCAGCAACAACAACGACUGCAAGGGAGAGAGAGGAAAAACAAAGAAAACUGCGGGGAAACAGUGAAAUUCAGUCA ..((((((((((((((((.....))))....---....((.(...(((((.........).))))...).)).))))))))......((....))))))(....)............... ( -24.70) >consensus UUCUGCUUCUUCCCGUGGAGAUGCUUCAACAUCAACGACUUCGAAGCAGCAACAACAAC_ACUGCACGGGAGCAAGAAGAAAAACCAAGA_AACUGCUG_GAAACAGUGAAAAUCGGUUA ((((.(((.((((((((.((.((((((...............))))))(......).....)).)))))))).))).)))).((((......((((........)))).......)))). (-26.68 = -27.30 + 0.62)

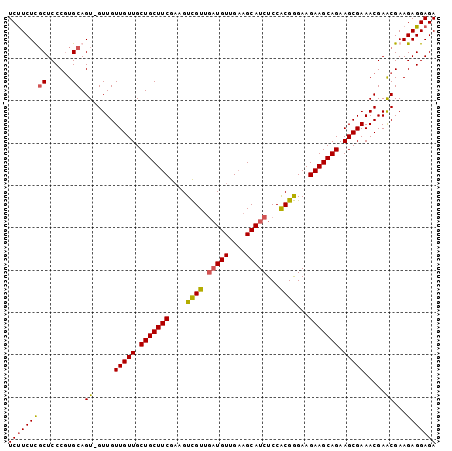
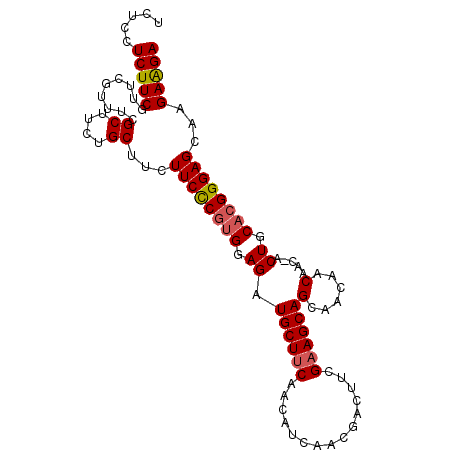
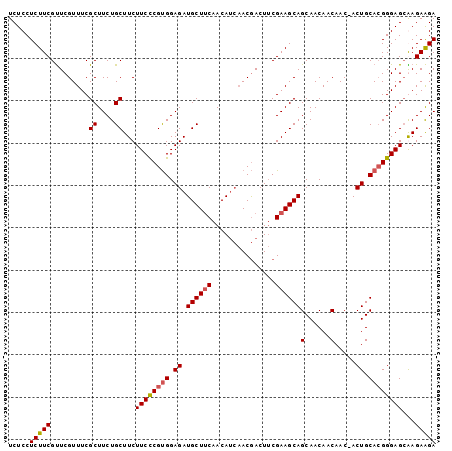
| Location | 1,551,782 – 1,551,882 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 92.87 |
| Mean single sequence MFE | -34.25 |
| Consensus MFE | -29.23 |
| Energy contribution | -29.23 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1551782 100 + 23771897 UCUUCUCGCUCCCGUGCAGU-GUUGUUGUUGCUGCUUCGAAGUCGUUGAUGUUGAAGCAUCUCCACGGGAAGAAGCAGAAGCGAAACGAACGAAGAGGAGA (((((((((......))..(-(((.(((((.(((((((....((((.(((((....)))))...))))...))))))).)))))))))......))))))) ( -34.60) >DroSec_CAF1 9490 100 + 1 UCUUCUUGCUCCCGUGCAGU-GUUGUUGUUGCUGCUUCGAAGUCGUUGAUGUUGAAGCAUCUCCACGGGAAGAAGCAGAAGCGAAACGAACGAAGAGGAGA (((((((((......))..(-(((.(((((.(((((((....((((.(((((....)))))...))))...))))))).)))))))))....))))))).. ( -32.50) >DroSim_CAF1 9609 100 + 1 UCUUCUUGCUCCCGUGCAGU-GUUGUUGUUGCUGCUUCGAAGUCGUUGAUGUUGAAGCAUCUCCACGGGAAGAAGCAGAAGCGAAACGAACGAAGAGGAGA (((((((((......))..(-(((.(((((.(((((((....((((.(((((....)))))...))))...))))))).)))))))))....))))))).. ( -32.50) >DroYak_CAF1 10324 98 + 1 UCCUCUCUCUCCCUUGCAGUCGUUGUUGUUGCUGCUUCGCAGUUGU---UGUUGCAGCAUCUCCGCGAGAAGAAGCAGAAGCGAAACGAGCGAAGAGGAGA ...((((.(((..((((..(((((.(((((.(((((((...(((((---....))))).((((...)))).))))))).)))))))))))))).))))))) ( -37.40) >consensus UCUUCUCGCUCCCGUGCAGU_GUUGUUGUUGCUGCUUCGAAGUCGUUGAUGUUGAAGCAUCUCCACGGGAAGAAGCAGAAGCGAAACGAACGAAGAGGAGA (((((((((......)).((.....(((((.(((((((....((((.(((((....)))))...))))...))))))).))))).....))...))))))) (-29.23 = -29.23 + -0.00)

| Location | 1,551,782 – 1,551,882 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 92.87 |
| Mean single sequence MFE | -27.80 |
| Consensus MFE | -22.53 |
| Energy contribution | -22.91 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.510804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
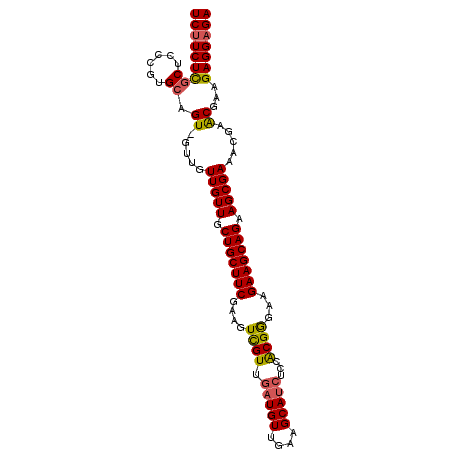
>3L_DroMel_CAF1 1551782 100 - 23771897 UCUCCUCUUCGUUCGUUUCGCUUCUGCUUCUUCCCGUGGAGAUGCUUCAACAUCAACGACUUCGAAGCAGCAACAACAAC-ACUGCACGGGAGCGAGAAGA .....(((((..((((...((....))....(((((((.((.((((((...............))))))(......)...-.)).)))))))))))))))) ( -27.96) >DroSec_CAF1 9490 100 - 1 UCUCCUCUUCGUUCGUUUCGCUUCUGCUUCUUCCCGUGGAGAUGCUUCAACAUCAACGACUUCGAAGCAGCAACAACAAC-ACUGCACGGGAGCAAGAAGA .....(((((.........((....))...((((((((.((.((((((...............))))))(......)...-.)).))))))))...))))) ( -26.16) >DroSim_CAF1 9609 100 - 1 UCUCCUCUUCGUUCGUUUCGCUUCUGCUUCUUCCCGUGGAGAUGCUUCAACAUCAACGACUUCGAAGCAGCAACAACAAC-ACUGCACGGGAGCAAGAAGA .....(((((.........((....))...((((((((.((.((((((...............))))))(......)...-.)).))))))))...))))) ( -26.16) >DroYak_CAF1 10324 98 - 1 UCUCCUCUUCGCUCGUUUCGCUUCUGCUUCUUCUCGCGGAGAUGCUGCAACA---ACAACUGCGAAGCAGCAACAACAACGACUGCAAGGGAGAGAGAGGA ((((((((((.((.((.(((.((((((........)))))).((((((..(.---........)..)))))).......)))..)).)))))))).)))). ( -30.90) >consensus UCUCCUCUUCGUUCGUUUCGCUUCUGCUUCUUCCCGUGGAGAUGCUUCAACAUCAACGACUUCGAAGCAGCAACAACAAC_ACUGCACGGGAGCAAGAAGA .....(((((.........((....))...((((((((.((.((((((...............))))))(......).....)).))))))))...))))) (-22.53 = -22.91 + 0.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:18 2006