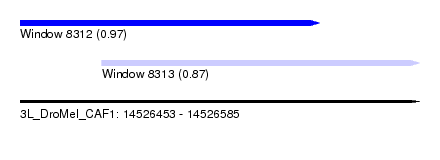
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,526,453 – 14,526,585 |
| Length | 132 |
| Max. P | 0.965189 |

| Location | 14,526,453 – 14,526,552 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 76.64 |
| Mean single sequence MFE | -25.34 |
| Consensus MFE | -15.32 |
| Energy contribution | -15.98 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14526453 99 + 23771897 UGCAACCCCUUUUCUCC--GC----UCGCCACU--------UGUACAAAUUGCUCGCUGAUUAGCAGGGAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUCGUCCACUUUUCUU ......((((((..((.--((----..((....--------..........))..)).))..)).))))..((((((((......)))))))).................... ( -17.84) >DroVir_CAF1 1090 101 + 1 UGCACCCCUUUUGGGGCAGCCUUGUCAGCCAGU--------UGUACAAAUUGCUCGCUGAUUAGCAGGCAAGCGCAUGAAAUUAAUCAUAUGCAAAUA----CGAGUAUGCUA .(((((((....))))..((((.((((((.(((--------.((....)).))).))))))....))))..(((.((((......)))).))).....----......))).. ( -30.90) >DroPse_CAF1 1027 107 + 1 -CUAACCCCUUUUUGCCUGCC----UUGCCACUCACUCAGUCGUACAAA-UGCUCGCUGAUUAGCAGGCAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUCGCCACUUUUUCCG -...........((((((((.----.((.....)).((((.((..(...-.)..))))))...))))))))((((((((......)))))))).................... ( -26.10) >DroGri_CAF1 1129 93 + 1 UGCAACCCCUUUCGG----------UUGGCAGU--------UGUACAAAUUGCUCGCUGAUUAGCAGGCAAGCGUAUGAAAUUAAUCAUAUGCAAAUAG--CCAAGUAUCCUU (((((((......))----------))(((...--------........((((..(((....)))..))))((((((((......)))))))).....)--))..)))..... ( -27.10) >DroAna_CAF1 977 99 + 1 GCUAACCCCUUUUUUGU--GC----UCGCCACU--------UGUACAAAUUGCUGGCUGAUUAGCAGGCAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUUGUCGACUUUUCUU (((((..((...(((((--((----........--------.))))))).....))....))))).(((((((((((((......)))))))).....))))).......... ( -24.00) >DroPer_CAF1 1027 107 + 1 -CUAACCCCUUUUUGCCUGCC----UUGCCACUCACUCAGUCGUACAAA-UGCUCGCUGAUUAGCAGGCAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUCGCCACUUUUUCCG -...........((((((((.----.((.....)).((((.((..(...-.)..))))))...))))))))((((((((......)))))))).................... ( -26.10) >consensus UCCAACCCCUUUUUGCC__CC____UUGCCACU________UGUACAAAUUGCUCGCUGAUUAGCAGGCAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUCGCCAACUUUUCUU .................................................((((..(((....)))..))))((((((((......)))))))).................... (-15.32 = -15.98 + 0.67)


| Location | 14,526,480 – 14,526,585 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.24 |
| Mean single sequence MFE | -28.52 |
| Consensus MFE | -17.06 |
| Energy contribution | -17.08 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.865047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14526480 105 + 23771897 UG-UACAAAUUGCUCGCUGAUUAGCAGGGAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUCGUCCACUUUUCUUUU-----CCUUGACGCG--AAUUUGUUGCUGUCGUGGGAG ..-.........(((((.(((.(((((.(((((((((((......))))))))......((((............-----....))))..--..))).))))))))))))).. ( -26.39) >DroGri_CAF1 1152 90 + 1 UG-UACAAAUUGCUCGCUGAUUAGCAGGCAAGCGUAUGAAAUUAAUCAUAUGCAAAUAG--CCAAGUAUCCUUCC-----AUUUGGCGCA--A-UGUGUCU------------ ..-((((..((((..(((....)))..))))((((((((......)))))))).....(--((((((........-----)))))))...--.-))))...------------ ( -27.60) >DroEre_CAF1 978 110 + 1 UG-GGCAAAUUGCUCGCUGAUUAGCAGGCAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUCGUCCACUUUUCUUUUCCUUUCCUUGAAGCG--AAUUUGUUGCUGUCGUGGGAG ((-(((...((((..(((....)))..))))((((((((......)))))))).......))))).............((((.(((((((--(.....))))).))).)))). ( -34.20) >DroWil_CAF1 35310 105 + 1 UGGGACAAAUUGCUCGCUGAUUAGCAAGCAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUUGCAAACUUUCUCUUU-----CUUUUUUUAU--AGCUUCUUUCUGUG-UCGCAA ((.((((..(((((.(((....))).)))))((((((((......))))))))......................-----..........--............))-)).)). ( -24.70) >DroYak_CAF1 984 105 + 1 UG-UGCAAAUUGCUCGCUGAUUAGCAGGCAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUCGUCCACUUUUCUUUU-----CCUUGACGCG--AAUUUGUUGCUGUCGUGGGAG .(-((....((((..(((....)))..))))((((((((......))))))))..........))).......((-----((.(((((((--(.....)))).)))).)))). ( -30.60) >DroAna_CAF1 1004 107 + 1 UG-UACAAAUUGCUGGCUGAUUAGCAGGCAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUUGUCGACUUUUCUUUU-----CCUUGACGCCCAAUGUUGUUGCUGUCGUGGGAG .(-.((((.(((((.(((....))).)))))((((((((......)))))))).....)))))..........((-----((.((((((.((....))..)).)))).)))). ( -27.60) >consensus UG_UACAAAUUGCUCGCUGAUUAGCAGGCAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUCGUCCACUUUUCUUUU_____CCUUGACGCG__AAUUUGUUGCUGUCGUGGGAG .........(((((.(((....))).)))))((((((((......))))))))............................................................ (-17.06 = -17.08 + 0.03)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:40 2006