
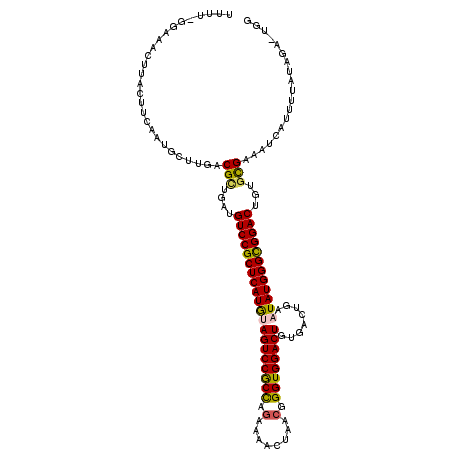
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,515,242 – 14,515,391 |
| Length | 149 |
| Max. P | 0.942784 |

| Location | 14,515,242 – 14,515,359 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.29 |
| Mean single sequence MFE | -37.11 |
| Consensus MFE | -32.08 |
| Energy contribution | -31.55 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14515242 117 + 23771897 UUUU-GUAAACUUACUUCAACGAUUGACGGGGAUGUCCGCUCAUGAAGUCCGCCAGAAAACUAACGGGUGGACUGUGACUGAAUAUGGGCGGACUGUGCGAAAUCAUUUUAUAUA-UGG ((((-(((........((..((.....))..)).(((((((((((.((((((((.(........).)))))))).........)))))))))))..)))))))............-... ( -35.40) >DroVir_CAF1 39786 117 + 1 UUU--UCAAACUUACUUUAAUGUUUGACGCUGAUGUCCGCUCAUGUAGUCCGCUAGGAAACUAACGGGUGGACUGUGACUGAAUAUGGGUGGACUGUGUGAAAUCAUUUUAUAGGCUGU ...--.....((((....(((((((.((((....((((((((((((((((((((((....))....))))))))........)))))))))))).)))).))).))))...)))).... ( -39.00) >DroGri_CAF1 14981 118 + 1 UUAUGUG-CACUUACUUCAAUGUUUGACGCUGACGUCCGCUCAUAUAGUCCACCAUGAAACUAAUGGGUGGACUGUGACUGAAUAUGGGUGGACUGUGUGAAAUCAUUUUAUAGAUUGC ......(-((.((.....(((((((.((((....((((((((((((((((((((((.......)).))))))))........)))))))))))).)))).))).)))).....)).))) ( -37.20) >DroWil_CAF1 14986 101 + 1 ---A-GGAAACUUACUUCAAUGCUUGACGCUGAUGUCCGCUCAUGAAGUCCGCCAGAAAACUAACGGGUGGACUGUGACUGAAUAUGGGCGGACUGUGUGAAAUC-------------- ---(-(....))...........((.((((....(((((((((((.((((((((.(........).)))))))).........))))))))))).)))).))...-------------- ( -35.40) >DroMoj_CAF1 31716 118 + 1 UUUUAUGAAACUUACUUCAGGGGUUGACGUUGAUGUCCGCUCAUAUAGUCCGCUAGGAAACUAACGGGUGGACUGUGACUGAAUAUGGGUGGACUGUGCGAAAUCAUUUUAUAGA-UGG .((((((((((((.....)))((((..(((....((((((((((((((((((((((....))....))))))))........))))))))))))...))).)))).)))))))))-... ( -38.80) >DroPer_CAF1 12321 117 + 1 UUGG-GUUAACUUACUUCAAUGCCGGCCGCUGAUGUCCGCUCAUGUAGUCCGCCAGAAAACUGACGGGUGGACUGUGACUGAAUAUGGGCGGACUGUGCGAAAUCAUUUUAUAUA-GAU ...(-((..............)))...(((....((((((((((((((((((((.(........).))))))))........))))))))))))...)))...............-... ( -36.84) >consensus UUUU_GGAAACUUACUUCAAUGCUUGACGCUGAUGUCCGCUCAUGUAGUCCGCCAGAAAACUAACGGGUGGACUGUGACUGAAUAUGGGCGGACUGUGCGAAAUCAUUUUAUAGA_UGG ...........................(((....((((((((((((((((((((.(........).))))))))........))))))))))))...)))................... (-32.08 = -31.55 + -0.53)


| Location | 14,515,281 – 14,515,391 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.00 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -19.80 |
| Energy contribution | -19.17 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14515281 110 + 23771897 UCAUGAAGUCCGCCAGAAAACUAACGGGUGGACUGUGACUGAAUAUGGGCGGACUGUGCGAAAUCAUU-UUAUAUA-UGGCUUCGAUCUGCG--------CUGAUUACAAUGAUCUCUAC ((((..((((((((.(........).))))))))))))........(.(((((.....((((..(((.-......)-))..)))).))))).--------).((((.....))))..... ( -28.50) >DroVir_CAF1 39824 98 + 1 UCAUGUAGUCCGCUAGGAAACUAACGGGUGGACUGUGACUGAAUAUGGGUGGACUGUGUGAAAUCAUU-UUAUAGGCUGUCGUUUCUCUA-------------------UAGAUCUCU-- (((..(((((((((((....)).((((.....))))...........)))))))))..)))......(-(((((((..(....)..))))-------------------)))).....-- ( -28.60) >DroPse_CAF1 11806 116 + 1 UCAUGUAGUCCGCCAGAAAACUGACGGGUGGACUGUGACUGAAUAUGGGCGGACUGUGCGAAAUCAUU-UUAUAUA-GAUCUGUGAUCUGCUGUUUCCUAUUGAUUAUAUUGAUCUCG-- ((.(..((((((((.......(.((((.....)))).).........))))))))..).)).......-......(-(((((((((((..............)))))))..)))))..-- ( -28.13) >DroGri_CAF1 15020 104 + 1 UCAUAUAGUCCACCAUGAAACUAAUGGGUGGACUGUGACUGAAUAUGGGUGGACUGUGUGAAAUCAUU-UUAUAGAUUGCUUUCUUUCUG-------------UAGGCGUAGAUCGCU-- ((((((((((((((.........((((.....))))...........)))))))))))))).......-.((((((..(....)..))))-------------))((((.....))))-- ( -31.55) >DroMoj_CAF1 31756 97 + 1 UCAUAUAGUCCGCUAGGAAACUAACGGGUGGACUGUGACUGAAUAUGGGUGGACUGUGCGAAAUCAUU-UUAUAGA-UGGAUUUUCUCUA-------------------UAGAUCUCU-- ((.(((((((((((((....)).((((.....))))...........))))))))))).))......(-(((((((-.((....))))))-------------------)))).....-- ( -27.20) >DroAna_CAF1 11518 111 + 1 UCAUGAAGUCCGCCAGAAAACUAACGGGUGGACUGUGACUGAAUAUGGGCGGACUGUGUGAAAUCAUUUUUAUAUU-GAUUUUCGAUAGGCG--------CUGAUAUCAGUGAUCUCUAC ......((((((((.(........).))))))))((.(((((.(((.((((..((((..(((((((.........)-))))))..)))).))--------)).)))))))).))...... ( -35.90) >consensus UCAUGUAGUCCGCCAGAAAACUAACGGGUGGACUGUGACUGAAUAUGGGCGGACUGUGCGAAAUCAUU_UUAUAGA_UGUCUUCGAUCUG_____________AU__CAUAGAUCUCU__ ((.(((((((((((.........((((.....))))...........))))))))))).))........................................................... (-19.80 = -19.17 + -0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:37 2006