
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,507,373 – 14,507,504 |
| Length | 131 |
| Max. P | 0.922707 |

| Location | 14,507,373 – 14,507,467 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 74.03 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -21.66 |
| Energy contribution | -21.38 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
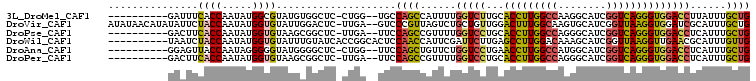
>3L_DroMel_CAF1 14507373 94 + 23771897 ----------GAUUUCACCAAUAUGGCGUAUGUGGCUC-CUGG--UGCCAGCCAUUUUGGUCUUGCACCUUGGCCAAGGCAUCGGUCAGGGUGGACCUUAUUUGCUG ----------......(((((.(((((.....((((..-....--.))))))))).)))))....((((((((((........)))))))))).............. ( -34.30) >DroVir_CAF1 32075 104 + 1 AUAUAACAUAUAUUCUACCAAUAUGGUGUAUUGGACUC-UUGA--GUCCCGUUAGUCUGCUGUUGGACUUUGGCAAGUGCAUCGGUUAAGGUGGAUCGCAUUUGCUG .............((((((....((((((((((((((.-...)--)))).((((((((......)))))..))).))))))))).....))))))..((....)).. ( -29.40) >DroPse_CAF1 3890 94 + 1 ----------GACUUCACCAAUAUGGUGUAAGCGGCUC-UUGA--UUCCAGCCGUUUUGGUCCUGCACCUUGGCCAGGGCAUCGGUCAGGGUGGACCUCAUUUGCUG ----------.....((((.....)))).((((((((.-....--....)))))))).(((((...(((((((((........)))))))))))))).......... ( -35.40) >DroWil_CAF1 5684 97 + 1 ----------UAAUCUACCAAUAUGGUGUAUUUGUAUCACCGGCACUCCAACCAUUCGAUUCUUGAGCCUUGGACAAAGCAUCGGUUAAGGUUGAACGCAUUUGUUG ----------........((((((((((.........)))))((....(((((..(((((.(((...(....)...))).)))))....)))))...))...))))) ( -20.10) >DroAna_CAF1 3786 94 + 1 ----------GGAGUUACCAAUAGGGGGUAUGGGGCUC-CUGG--UUCCAGCUGUUCUGGUCCUGAACCUUGGCCAUGGCAUCGGUCAGGGUGGACCUCAUUUGCUG ----------(((...((((....(((((.....))))-))))--))))(((......(((((...(((((((((........))))))))))))))......))). ( -36.80) >DroPer_CAF1 4415 94 + 1 ----------GACUUCACCAAUAUGGUGUAAGCGGCUC-UUGA--UUCCAGCCGUUUUGGUCCUGCACCUUGGCCAGGGCAUCGGUCAGGGUGGACCUCAUUUGCUG ----------.....((((.....)))).((((((((.-....--....)))))))).(((((...(((((((((........)))))))))))))).......... ( -35.40) >consensus __________GAAUUCACCAAUAUGGUGUAUGCGGCUC_CUGA__UUCCAGCCAUUCUGGUCCUGCACCUUGGCCAGGGCAUCGGUCAGGGUGGACCUCAUUUGCUG ...............((((.....))))....................((((......(((((...(((((((((........))))))))))))))......)))) (-21.66 = -21.38 + -0.27)


| Location | 14,507,390 – 14,507,504 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 77.80 |
| Mean single sequence MFE | -40.22 |
| Consensus MFE | -23.34 |
| Energy contribution | -23.65 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14507390 114 + 23771897 GUAUGUGGCUC-CUGG--UGCCAGCCAUUUUGGUCUUGCACCUUGGCCAAGGCAUCGGUCAGGGUGGACCUUAUUUGCUGGCUCGAGUUGCGAUUCUGACCCUUAUCAUAGGGCGAC ...((..((((-...(--.((((((......((((...((((((((((........))))))))))))))......)))))).)))))..))....((.((((......)))))).. ( -48.10) >DroVir_CAF1 32102 114 + 1 GUAUUGGACUC-UUGA--GUCCCGUUAGUCUGCUGUUGGACUUUGGCAAGUGCAUCGGUUAAGGUGGAUCGCAUUUGCUGGCUGGAGUUACGGGUUUGACCCUUAUCAUAGGGCGAC ...(..(((((-.(((--.(((.((((((((......)))))..((((((((((((.(......).))).)))))))))))).))).))).)))))..)((((......)))).... ( -39.10) >DroPse_CAF1 3907 114 + 1 GUAAGCGGCUC-UUGA--UUCCAGCCGUUUUGGUCCUGCACCUUGGCCAGGGCAUCGGUCAGGGUGGACCUCAUUUGCUGACUGGAGUUGCGAUUCUGACCCUUAUCAUAGGGAGAG ....(((((((-(...--...((((......(((((...(((((((((........))))))))))))))......))))...))))))))...(((..((((......))))))). ( -44.80) >DroWil_CAF1 5701 117 + 1 GUAUUUGUAUCACCGGCACUCCAACCAUUCGAUUCUUGAGCCUUGGACAAAGCAUCGGUUAAGGUUGAACGCAUUUGUUGACUGGAGUUACGAUUUUGUCCCUUAUCAUAGGGCGAC ....(((((.(.(((((....(((((..(((((.(((...(....)...))).)))))....)))))((((....))))).)))).).)))))..(((.((((......))))))). ( -27.90) >DroMoj_CAF1 23972 111 + 1 GUAUUUGGUUC-CUGG--GUCCAGUUGUUCU---GUUGUGCUUUGGCAAGUGCAUCGGUUAAAGUGGAUCGCAUUUGCUGGCUGGAACUACGAUUUUGACCCUUAUCAUAGGGCGAC .....((((((-(..(--(((((.((...((---(.((..(((....)))..)).)))...)).))))))((....)).....))))))).....(((.((((......))))))). ( -36.20) >DroAna_CAF1 3803 114 + 1 GUAUGGGGCUC-CUGG--UUCCAGCUGUUCUGGUCCUGAACCUUGGCCAUGGCAUCGGUCAGGGUGGACCUCAUUUGCUGGCUAGAGUUGCGAUUCUGACCCUUAUCAUAGGGCGAC ...((.(((((-.(((--..(((((......(((((...(((((((((........))))))))))))))......))))))))))))).))....((.((((......)))))).. ( -45.20) >consensus GUAUGUGGCUC_CUGG__UUCCAGCCAUUCUGGUCUUGAACCUUGGCCAGGGCAUCGGUCAAGGUGGACCGCAUUUGCUGGCUGGAGUUACGAUUCUGACCCUUAUCAUAGGGCGAC ......(((((...(.....(((((......(((((...(((((((((........))))))))))))))......))))))..)))))..........((((......)))).... (-23.34 = -23.65 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:30 2006