
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,467,432 – 14,467,528 |
| Length | 96 |
| Max. P | 0.815690 |

| Location | 14,467,432 – 14,467,528 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.49 |
| Mean single sequence MFE | -28.74 |
| Consensus MFE | -22.62 |
| Energy contribution | -22.94 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
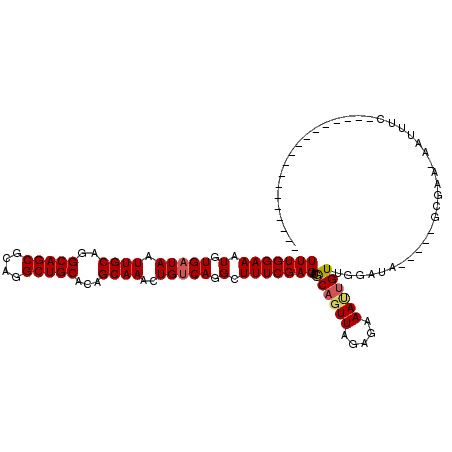
>3L_DroMel_CAF1 14467432 96 + 23771897 UUUGGAAAUGUGAUAAUUGCAGGCAGCGCAGGCUGCACAGCAAACUGUCAGGCUUUCGAGAUGCAGUUAGAGAAAAUGUUGAAUA-----GCGGA-AAUUUC------------------ ....(((((.(((((.((((..(((((....)))))...))))..))))).((((((((.((.............)).))))).)-----))...-.)))))------------------ ( -23.52) >DroSec_CAF1 64349 96 + 1 UUUGGAAAUGUGAUAAUUGCAGGCAGCGCAGGCUGCACAGCAAACUGUCAGGCUUUCGAGAUACAGUUAGAGAAAUUGUUGUAUA-----GCGGA-CAUUUC------------------ ....(((((((((((.((((..(((((....)))))...))))..)))))....((((..((((((...((....)).)))))).-----.))))-))))))------------------ ( -28.60) >DroSim_CAF1 79579 96 + 1 UUUGGAAAUGUGAUAAUUGCAGGCAGCGCAGGCUGCACAGCAAACUGUCAGGCUUUCGAGAUGCAGUUAGCGAAAUUGUUGGGUA-----GCAAG-AAUUUC------------------ (((((((.(.(((((.((((..(((((....)))))...))))..))))).).)))))))..((.....))((((((.(((....-----.))).-))))))------------------ ( -28.10) >DroEre_CAF1 80185 98 + 1 UUUGGAAAUGUGAUAAUUGCAGGCAGCGCAGGCUGCACAGCAAGCUGCCAGGCUUUCGAGAUGCAGUUAGAGAAACUGUUGGCAA-----GCGCCUAAUUUUU----------------- (((((((.((..(...)..))((((((....(((....)))..))))))....)))))))..((((((.....)))))).(((..-----..)))........----------------- ( -28.30) >DroYak_CAF1 71345 120 + 1 UUUGGAAAUGUGAUAAUUGCAGGCAGCGCAGGCUGCACAGCAAGCUGUCAGGCUUUCGAGAUGCAGUUAGAGAAAUGGUUGGAUAUGUUAGCUAACCAUCUUUAAAACCUAUUAGCAAGU (((((((.(.(((((.((((..(((((....)))))...))))..))))).).))))))).(((..(((((((..(((((((.........)))))))))))))).........)))... ( -35.20) >consensus UUUGGAAAUGUGAUAAUUGCAGGCAGCGCAGGCUGCACAGCAAACUGUCAGGCUUUCGAGAUGCAGUUAGAGAAAUUGUUGGAUA_____GCGAA_AAUUUC__________________ (((((((.(.(((((.((((..(((((....)))))...))))..))))).).)))))))..((((((.....))))))......................................... (-22.62 = -22.94 + 0.32)


| Location | 14,467,432 – 14,467,528 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.49 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -15.48 |
| Energy contribution | -15.32 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14467432 96 - 23771897 ------------------GAAAUU-UCCGC-----UAUUCAACAUUUUCUCUAACUGCAUCUCGAAAGCCUGACAGUUUGCUGUGCAGCCUGCGCUGCCUGCAAUUAUCACAUUUCCAAA ------------------(((((.-.....-----.................(((((((...(....)..)).)))))(((.(.(((((....)))))).)))........))))).... ( -18.20) >DroSec_CAF1 64349 96 - 1 ------------------GAAAUG-UCCGC-----UAUACAACAAUUUCUCUAACUGUAUCUCGAAAGCCUGACAGUUUGCUGUGCAGCCUGCGCUGCCUGCAAUUAUCACAUUUCCAAA ------------------((((((-(....-----.................((((((....(....)....))))))(((.(.(((((....)))))).)))......))))))).... ( -24.10) >DroSim_CAF1 79579 96 - 1 ------------------GAAAUU-CUUGC-----UACCCAACAAUUUCGCUAACUGCAUCUCGAAAGCCUGACAGUUUGCUGUGCAGCCUGCGCUGCCUGCAAUUAUCACAUUUCCAAA ------------------((((((-.(((.-----....))).))))))((.(((((((...(....)..)).))))).))((((((((....)))))..)))................. ( -21.70) >DroEre_CAF1 80185 98 - 1 -----------------AAAAAUUAGGCGC-----UUGCCAACAGUUUCUCUAACUGCAUCUCGAAAGCCUGGCAGCUUGCUGUGCAGCCUGCGCUGCCUGCAAUUAUCACAUUUCCAAA -----------------........((.((-----(.((((.(((((.....))))).....(....)..)))))))((((.(.(((((....)))))).))))...........))... ( -26.50) >DroYak_CAF1 71345 120 - 1 ACUUGCUAAUAGGUUUUAAAGAUGGUUAGCUAACAUAUCCAACCAUUUCUCUAACUGCAUCUCGAAAGCCUGACAGCUUGCUGUGCAGCCUGCGCUGCCUGCAAUUAUCACAUUUCCAAA ....(((..((((((((..((((((((((.....................)))))..)))))..))))))))..)))((((.(.(((((....)))))).))))................ ( -30.50) >consensus __________________GAAAUU_UCCGC_____UAUCCAACAAUUUCUCUAACUGCAUCUCGAAAGCCUGACAGUUUGCUGUGCAGCCUGCGCUGCCUGCAAUUAUCACAUUUCCAAA ...............................................................((((...(((.((.((((.(.(((((....)))))).)))))).)))..)))).... (-15.48 = -15.32 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:18 2006