| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
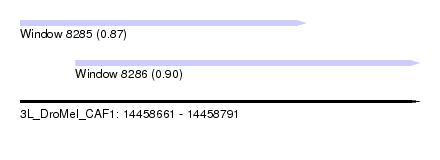
| Location | 14,458,661 – 14,458,791 |
| Length | 130 |
| Max. P | 0.898323 |

| Location | 14,458,661 – 14,458,754 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 79.62 |
| Mean single sequence MFE | -25.10 |
| Consensus MFE | -13.03 |
| Energy contribution | -14.20 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
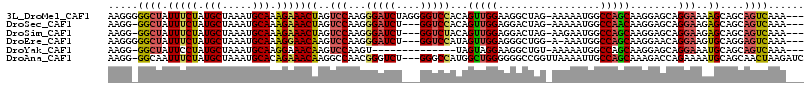
>3L_DroMel_CAF1 14458661 93 + 23771897 ACAGAAGUUUACUUGGUUAAGGGGGCUAUUUCUAUGCUAAAUGCAAAGAAACUAGUCCAAGGGAUCUAGGGGUCCACAGUUGGAAGGCUAG-AA .....(((((.(((((((....(((((((((((.(((.....))).))))).))))))....)).)))))..((((....)))))))))..-.. ( -26.00) >DroSec_CAF1 55700 89 + 1 ACAGAAGUUUACUUGGUUAAGG-GGCUAUUUCUAUGCUAAAUGCAAAGAAACUAGUCCAAGGGAUCU---GGUCCACAGUUGGAGGACUAG-AA .....................(-((((((((((.(((.....))).))))).))))))......(((---(((((.(.....).)))))))-). ( -27.70) >DroSim_CAF1 70870 89 + 1 ACAGAAGUUUACUUGGUUAAGG-GGCUAUUUCUAUGCUAAAUGCAAAGAAACUAGUCCAAGGGAUCU---GGUCUACAGUUGGAGGACUAG-AA .....................(-((((((((((.(((.....))).))))).))))))......(((---(((((.(.....).)))))))-). ( -25.80) >DroEre_CAF1 71316 89 + 1 ACAGAAGUUUACUUGGUUAAGGGGGCUAUUUCUAUGCUAAAUGCAAAGGAACAAGUCCAAGGGAUCU---GGUCCAUAGUUGGAGGGCUGG-A- .(((.......((.((((....(((((.(((((.(((.....))).)))))..)))))....)))).---))((((....))))...))).-.- ( -23.80) >DroYak_CAF1 62172 78 + 1 ACAGAAGUUUACUUGGUUAAGG-GGCUAUUCCUAUGCUAAAUGCAAGGAAACAAGUCCAAGU--------------UAGUAGGAAGGCUGU-AA ((((...((((((........(-((((.(((((.(((.....))))))))...)))))....--------------.))))))....))))-.. ( -21.92) >DroAna_CAF1 68997 79 + 1 ACAGAAA-----------AAGG-GGCAAUUUCUAUGCUAAAUGCACAGAAACAAGGCCAACGGGUCU---GGGCCAUGGCUGGGGGGCCGGUUA .((((..-----------....-(((..(((((.(((.....))).)))))....)))......)))---)((((..((((....)))))))). ( -25.40) >consensus ACAGAAGUUUACUUGGUUAAGG_GGCUAUUUCUAUGCUAAAUGCAAAGAAACAAGUCCAAGGGAUCU___GGUCCACAGUUGGAGGGCUAG_AA .......................((((.(((((.(((.....))).)))))....(((((.(((((....)))))....))))).))))..... (-13.03 = -14.20 + 1.17)

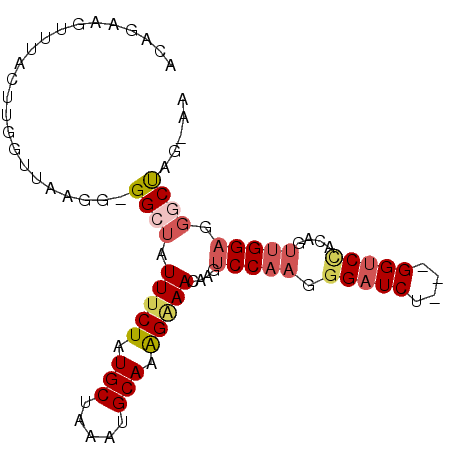
| Location | 14,458,679 – 14,458,791 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 81.37 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -19.88 |
| Energy contribution | -19.96 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14458679 112 + 23771897 AAGGGGGCUAUUUCUAUGCUAAAUGCAAAGAAACUAGUCCAAGGGAUCUAGGGGUCCACAGUUGGAAGGCUAG-AAAAAUGGCCAGCAAGGAGCAGGAAAAGCAGCAGUCAAA--- ....(((((((((((.(((.....))).))))).))))))...(((((....)))))...((((...(((((.-.....))))).((.....))........)))).......--- ( -35.30) >DroSec_CAF1 55718 108 + 1 AAGG-GGCUAUUUCUAUGCUAAAUGCAAAGAAACUAGUCCAAGGGAUCU---GGUCCACAGUUGGAGGACUAG-AAAAAUGGCCAACAAGGAGCAGGAAGAGCAGCAGUCAAA--- ...(-((((((((((.(((.....))).))))).))))))......(((---(.(((...(((((....(((.-.....))))))))..))).))))................--- ( -30.60) >DroSim_CAF1 70888 108 + 1 AAGG-GGCUAUUUCUAUGCUAAAUGCAAAGAAACUAGUCCAAGGGAUCU---GGUCUACAGUUGGAGGACUAG-AAGAAUGGCCAGCAAGGAGCAGGAAGAGCAGCAGUCAAA--- ...(-((((((((((.(((.....))).))))).))))))......(((---(((((.(.....).)))))))-)....((((..((...(..(.....)..).)).))))..--- ( -29.40) >DroEre_CAF1 71334 108 + 1 AAGGGGGCUAUUUCUAUGCUAAAUGCAAAGGAACAAGUCCAAGGGAUCU---GGUCCAUAGUUGGAGGGCUGG-A-AAAUGGCCAGCAAGGAACAGGAAGUGCAGGAGUCAAA--- ....(((((.(((((.(((.....))).)))))..)))))..((..(((---(.((((....))))..(((((-.-......)))))...............))))..))...--- ( -29.20) >DroYak_CAF1 62190 97 + 1 AAGG-GGCUAUUCCUAUGCUAAAUGCAAGGAAACAAGUCCAAGU--------------UAGUAGGAAGGCUGU-AAAAAUGGCCAGCAAGGAGCAGGAAAUGCAGCAGUCAAA--- ...(-((((.(((((.(((.....))))))))...)))))....--------------......((..(((((-(.......((.((.....)).))...))))))..))...--- ( -27.50) >DroAna_CAF1 69004 112 + 1 AAGG-GGCAAUUUCUAUGCUAAAUGCACAGAAACAAGGCCAACGGGUCU---GGGCCAUGGCUGGGGGGCCGGUUAAAAUUGCCAGCAAAGACCAGAAAAUGCAGCAACUAAGAUC .((.-.((..(((((.(((.....))).)))))...((((..(.((((.---.......)))).)..))))((((....(((....))).))))..........))..))...... ( -32.20) >consensus AAGG_GGCUAUUUCUAUGCUAAAUGCAAAGAAACAAGUCCAAGGGAUCU___GGUCCACAGUUGGAGGGCUAG_AAAAAUGGCCAGCAAGGAGCAGGAAAAGCAGCAGUCAAA___ .....((((.(((((.(((.....))).)))))((..(((...(((((....)))))...(((((.................)))))........)))..))....))))...... (-19.88 = -19.96 + 0.09)
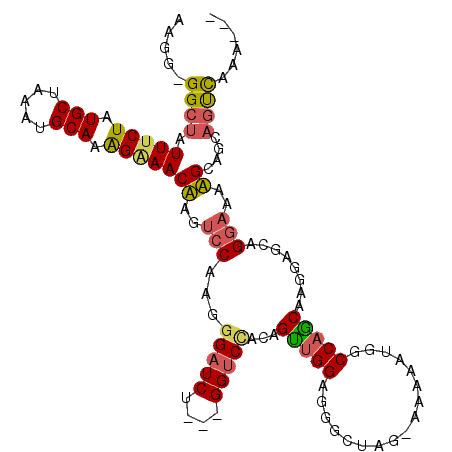
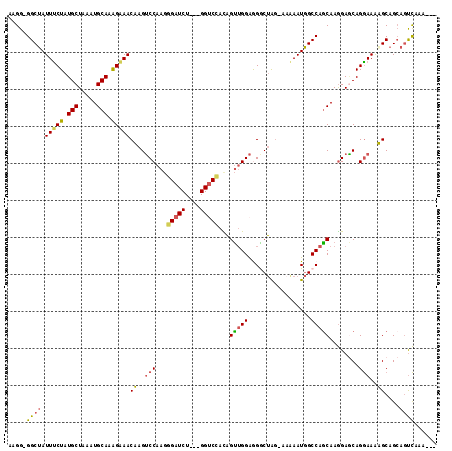
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:15 2006