
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,425,989 – 14,426,176 |
| Length | 187 |
| Max. P | 0.998602 |

| Location | 14,425,989 – 14,426,081 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.40 |
| Mean single sequence MFE | -20.80 |
| Consensus MFE | -12.13 |
| Energy contribution | -13.30 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14425989 92 - 23771897 --GAAAAGACCAAACCUAAAGCCAAUAAGUGGCAUUAGCCAAAUGUGGU-AAAACCAUUUGGCAACUUGUUAAUAAGCA-AUUGCAC----------------------AGCA-CAAAU --...(((............((((.....))))....((((((((.(..-....)))))))))..)))........((.-...))..----------------------....-..... ( -19.60) >DroPse_CAF1 49223 116 - 1 --AGAAAGACCUAGCCUAAAGCCAAUAAAUGGCAUUAGCCAAAUGCGGUUAAGACAAUUUUGAAACUUGUUAAUAAGCG-AUUGCACUCACACGAUCGGCAUCAGCACCAGCACCCAAG --...........(((....((((.....)))).((((((......))))))(((((.........))))).......(-((((........))))))))....((....))....... ( -21.20) >DroSec_CAF1 28342 92 - 1 --GAAAAGACCAAACCUAAAGCCAAUAAGUGGCAUUGGCCAAAUGUGGU-AAAACCAUUUGGCAACUUGUUAAUAAGCA-AUUGCAC----------------------AGCA-CAAAU --..................((((.....)))).(((((((((((.(..-....)))))))((((.(((((....))))-)))))..----------------------.)).-))).. ( -19.90) >DroYak_CAF1 33838 94 - 1 GCGAAAAGACCAAACCUAAAGCCAAUAAGUGGCAUUAGCCAAAUGUGGU-AAAACCAUUUGGCAACUUGUUAAUAAGCA-AUUGCAC----------------------AGCA-CAAAU ((((................((((.....))))....((((((((.(..-....)))))))))...(((((....))))-)))))..----------------------....-..... ( -22.20) >DroAna_CAF1 43570 91 - 1 --GAAAAGACCUAACCUAAAGCCAAUAAGUGGCAUUAGCCAAAUGUGGC-AAAACCAUUUGGCAACUUGUUAAUAAGCAAAUUGCAU----------------------A--A-ACAAU --..................((((.....((((....))))....))))-...........((((.(((((....))))).))))..----------------------.--.-..... ( -20.70) >DroPer_CAF1 49389 116 - 1 --AGAAAGACCUAGCCUAAAGCCAAUAAAUGGCAUUAGCCAAAUGCGGUUAAGACAAUUUUUAAACUUGUUAAUAAGCG-AUUGCACUCACACGAUCGGCAUCAGCACCAGCACCCAAG --...........(((....((((.....)))).((((((......))))))(((((.........))))).......(-((((........))))))))....((....))....... ( -21.20) >consensus __GAAAAGACCAAACCUAAAGCCAAUAAGUGGCAUUAGCCAAAUGUGGU_AAAACCAUUUGGCAACUUGUUAAUAAGCA_AUUGCAC______________________AGCA_CAAAU .....(((............((((.....))))....((((((((.(.......)))))))))..)))........((.....)).................................. (-12.13 = -13.30 + 1.17)



| Location | 14,426,005 – 14,426,100 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 79.75 |
| Mean single sequence MFE | -17.79 |
| Consensus MFE | -12.65 |
| Energy contribution | -13.60 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.839639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14426005 95 + 23771897 UGCUUAUUAACAAGUUGCCAAAUGGUUUU-ACCACAUUUGGCUAAUGCCACUUAUUGGCUUUAGGUUUGGUCUUUUCGU-UUU-UCAUUUUGCUAUUU .((.....((((((..(((((((((....-.))........((((.((((.....)))).)))))))))))..))).))-)..-.......))..... ( -18.44) >DroPse_CAF1 49262 78 + 1 CGCUUAUUAACAAGUUUCAAAAUUGUCUUAACCGCAUUUGGCUAAUGCCAUUUAUUGGCUUUAGGCUAGGUCUUUCUG-------------------- .((......((((.........)))).......))((((((((...((((.....))))....)))))))).......-------------------- ( -13.92) >DroSec_CAF1 28358 96 + 1 UGCUUAUUAACAAGUUGCCAAAUGGUUUU-ACCACAUUUGGCCAAUGCCACUUAUUGGCUUUAGGUUUGGUCUUUUCCUUUUU-UCAUUUUGCUAUUU .(((((.......(((((((((((.....-....))))))).))))((((.....))))..)))))..((......)).....-.............. ( -18.10) >DroEre_CAF1 43582 95 + 1 UGCUUAUUAACAAGUUGCCAAAUGGUUUU-ACCACAUUUGGCUAAUGCCACUUAUUGGCUUUAGGUUUGGUCUUUUCGU-UUU-UCAUUUUGCUAUUU .((.....((((((..(((((((((....-.))........((((.((((.....)))).)))))))))))..))).))-)..-.......))..... ( -18.44) >DroAna_CAF1 43585 94 + 1 UGCUUAUUAACAAGUUGCCAAAUGGUUUU-GCCACAUUUGGCUAAUGCCACUUAUUGGCUUUAGGUUAGGUCUUUUCGU-UUUCUUGUUUUAGC--UU .(((....((((((..((((((((.....-....))))))))(((.((((.....)))).)))................-...))))))..)))--.. ( -23.90) >DroPer_CAF1 49428 78 + 1 CGCUUAUUAACAAGUUUAAAAAUUGUCUUAACCGCAUUUGGCUAAUGCCAUUUAUUGGCUUUAGGCUAGGUCUUUCUG-------------------- .((......((((.........)))).......))((((((((...((((.....))))....)))))))).......-------------------- ( -13.92) >consensus UGCUUAUUAACAAGUUGCCAAAUGGUUUU_ACCACAUUUGGCUAAUGCCACUUAUUGGCUUUAGGUUAGGUCUUUUCGU_UUU_UCAUUUUGCU__UU .(((((..........((((((((..........))))))))....((((.....))))..)))))................................ (-12.65 = -13.60 + 0.95)



| Location | 14,426,005 – 14,426,100 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 79.75 |
| Mean single sequence MFE | -19.35 |
| Consensus MFE | -10.87 |
| Energy contribution | -12.03 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973704 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14426005 95 - 23771897 AAAUAGCAAAAUGA-AAA-ACGAAAAGACCAAACCUAAAGCCAAUAAGUGGCAUUAGCCAAAUGUGGU-AAAACCAUUUGGCAACUUGUUAAUAAGCA ...((((((.....-...-....................((((.....))))....((((((((.(..-....)))))))))...))))))....... ( -21.40) >DroPse_CAF1 49262 78 - 1 --------------------CAGAAAGACCUAGCCUAAAGCCAAUAAAUGGCAUUAGCCAAAUGCGGUUAAGACAAUUUUGAAACUUGUUAAUAAGCG --------------------............((.....((((.....)))).((((((......))))))(((((.........))))).....)). ( -12.90) >DroSec_CAF1 28358 96 - 1 AAAUAGCAAAAUGA-AAAAAGGAAAAGACCAAACCUAAAGCCAAUAAGUGGCAUUGGCCAAAUGUGGU-AAAACCAUUUGGCAACUUGUUAAUAAGCA ...((((((.....-.....((......)).........((((.....))))....((((((((.(..-....)))))))))...))))))....... ( -21.70) >DroEre_CAF1 43582 95 - 1 AAAUAGCAAAAUGA-AAA-ACGAAAAGACCAAACCUAAAGCCAAUAAGUGGCAUUAGCCAAAUGUGGU-AAAACCAUUUGGCAACUUGUUAAUAAGCA ...((((((.....-...-....................((((.....))))....((((((((.(..-....)))))))))...))))))....... ( -21.40) >DroAna_CAF1 43585 94 - 1 AA--GCUAAAACAAGAAA-ACGAAAAGACCUAACCUAAAGCCAAUAAGUGGCAUUAGCCAAAUGUGGC-AAAACCAUUUGGCAACUUGUUAAUAAGCA ..--(((..((((((...-....................((((.....))))....((((((((.(..-....)))))))))..))))))....))). ( -25.80) >DroPer_CAF1 49428 78 - 1 --------------------CAGAAAGACCUAGCCUAAAGCCAAUAAAUGGCAUUAGCCAAAUGCGGUUAAGACAAUUUUUAAACUUGUUAAUAAGCG --------------------............((.....((((.....)))).((((((......))))))(((((.........))))).....)). ( -12.90) >consensus AA__AGCAAAAUGA_AAA_ACGAAAAGACCAAACCUAAAGCCAAUAAGUGGCAUUAGCCAAAUGUGGU_AAAACCAUUUGGCAACUUGUUAAUAAGCA ........................(((............((((.....))))....((((((((.(.......)))))))))..)))........... (-10.87 = -12.03 + 1.17)

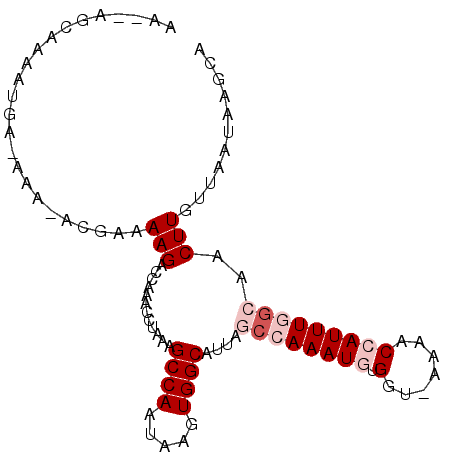

| Location | 14,426,081 – 14,426,176 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 91.37 |
| Mean single sequence MFE | -21.96 |
| Consensus MFE | -18.26 |
| Energy contribution | -19.14 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14426081 95 + 23771897 GU-UUU-UCAUUUUGCUAUUUGUAUUU-UAUCUCUUUUUCCAUAUUUCCAAUUGGAUGUCAGUGUGGGCUGUCGUCACGCCCACAAUCUCGGUAGCAG ..-...-......(((((((.......-..........((((..........))))......(((((((((....)).))))))).....))))))). ( -22.50) >DroSec_CAF1 28434 96 + 1 CUUUUU-UCAUUUUGCUAUUUGUAUUU-UAUCUCUUUUUCCAUAUUUCCAAUUGGAUGUCAGUGUGGGCUGUCGUCACGCCCACAAUCUCGGUAGCAG ......-......(((((((.......-..........((((..........))))......(((((((((....)).))))))).....))))))). ( -22.50) >DroSim_CAF1 43238 95 + 1 GU-UUU-UCAUUUUGCUAUUUGUAUUU-UAUCUCUUUUUCCAUAUUUCCAAUUGGAUGUCAGUGUGGGCUGUCGUCACGCCCACAAUCUCGGUAGCAG ..-...-......(((((((.......-..........((((..........))))......(((((((((....)).))))))).....))))))). ( -22.50) >DroEre_CAF1 43658 94 + 1 GU-UUU-UCAUUUUGCUAUUUGU-UUU-UAUCUCUUUUUCCAUAUUUCCAAUUGGAUGUCAGUGUGGGCUGUCGUCACGCCCACAAUCUCGGUAGCAG ..-...-......(((((((...-...-..........((((..........))))......(((((((((....)).))))))).....))))))). ( -22.50) >DroAna_CAF1 43661 95 + 1 GU-UUUCUUGUUUUAGC--UUGCGUUUUCUUAUAUUUUUUCAUAUUUCCAAUUGGAUGUCAGCGUGGGCUGUCGUCACGCCCACAAUCUCAGUAGCAG ..-.....((((...((--(.((((((....((((......))))........)))))).)))((((((((....)).)))))).........)))). ( -19.80) >consensus GU_UUU_UCAUUUUGCUAUUUGUAUUU_UAUCUCUUUUUCCAUAUUUCCAAUUGGAUGUCAGUGUGGGCUGUCGUCACGCCCACAAUCUCGGUAGCAG .............(((((((..................((((..........))))......(((((((((....)).))))))).....))))))). (-18.26 = -19.14 + 0.88)


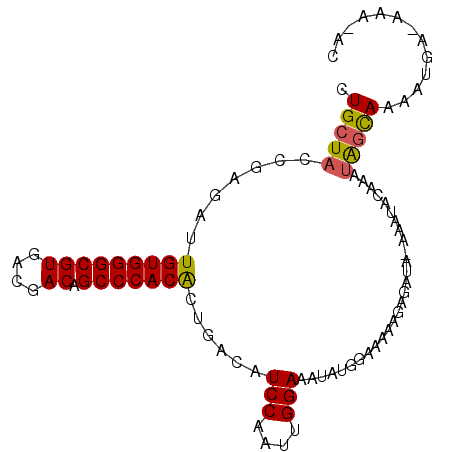
| Location | 14,426,081 – 14,426,176 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 91.37 |
| Mean single sequence MFE | -20.34 |
| Consensus MFE | -18.20 |
| Energy contribution | -18.12 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.998602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14426081 95 - 23771897 CUGCUACCGAGAUUGUGGGCGUGACGACAGCCCACACUGACAUCCAAUUGGAAAUAUGGAAAAAGAGAUA-AAAUACAAAUAGCAAAAUGA-AAA-AC .(((((.......(((((((((....)).)))))))((....((((..(....)..))))...)).....-.........)))))......-...-.. ( -20.60) >DroSec_CAF1 28434 96 - 1 CUGCUACCGAGAUUGUGGGCGUGACGACAGCCCACACUGACAUCCAAUUGGAAAUAUGGAAAAAGAGAUA-AAAUACAAAUAGCAAAAUGA-AAAAAG .(((((.......(((((((((....)).)))))))((....((((..(....)..))))...)).....-.........)))))......-...... ( -20.60) >DroSim_CAF1 43238 95 - 1 CUGCUACCGAGAUUGUGGGCGUGACGACAGCCCACACUGACAUCCAAUUGGAAAUAUGGAAAAAGAGAUA-AAAUACAAAUAGCAAAAUGA-AAA-AC .(((((.......(((((((((....)).)))))))((....((((..(....)..))))...)).....-.........)))))......-...-.. ( -20.60) >DroEre_CAF1 43658 94 - 1 CUGCUACCGAGAUUGUGGGCGUGACGACAGCCCACACUGACAUCCAAUUGGAAAUAUGGAAAAAGAGAUA-AAA-ACAAAUAGCAAAAUGA-AAA-AC .(((((.......(((((((((....)).)))))))((....((((..(....)..))))...)).....-...-.....)))))......-...-.. ( -20.60) >DroAna_CAF1 43661 95 - 1 CUGCUACUGAGAUUGUGGGCGUGACGACAGCCCACGCUGACAUCCAAUUGGAAAUAUGAAAAAAUAUAAGAAAACGCAA--GCUAAAACAAGAAA-AC ..(((.........((((((((....)).))))))((.....(((....))).((((......))))........)).)--))............-.. ( -19.30) >consensus CUGCUACCGAGAUUGUGGGCGUGACGACAGCCCACACUGACAUCCAAUUGGAAAUAUGGAAAAAGAGAUA_AAAUACAAAUAGCAAAAUGA_AAA_AC .(((((.......(((((((((....)).)))))))......(((....)))............................)))))............. (-18.20 = -18.12 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:06 2006