
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,425,381 – 14,425,477 |
| Length | 96 |
| Max. P | 0.654206 |

| Location | 14,425,381 – 14,425,477 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.85 |
| Mean single sequence MFE | -30.16 |
| Consensus MFE | -20.81 |
| Energy contribution | -21.83 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
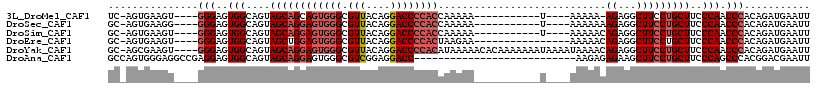
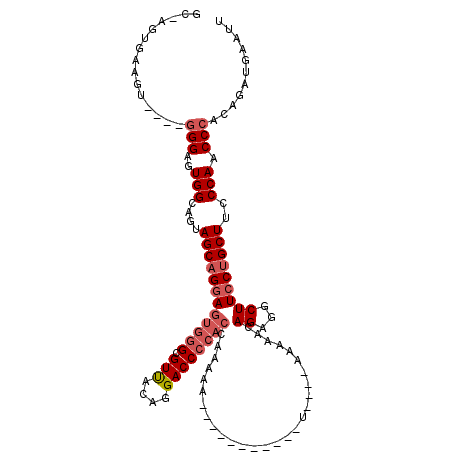
>3L_DroMel_CAF1 14425381 96 + 23771897 UC-AGUGAAGU----GGGAGUGGCAGUAGCAGCAGUGGGCGUUACAGGACCCCACCAAAAA-----------U----AAAAA-AGAGGCUUCCUGCUUCCCAACCCACAGAUGAAUU ..-.(((...(----((((..(((((.(((....(((((.(((....))))))))......-----------.----.....-....)))..))))))))))...)))......... ( -28.35) >DroSec_CAF1 27738 97 + 1 GC-AGUGAAGG----GGGAGUGGCAGUAGCAGGAGUGGGCGUUACAGGACCCCACCAAAAA-----------U----AAAAAAAGAGGCUUCCUGCUUCCCAACCCACAGAUGAAUU .(-(......(----((...(((....((((((((((((.(((....))))))))......-----------.----.............)))))))..))).))).....)).... ( -27.75) >DroSim_CAF1 42542 97 + 1 GC-AGUGAAGU----GGGAGUGGCAGUAGCAGGAGUGGGCGUUACAGGACCCCACCAAAAA-----------U----AAAAACAGAGGCUUCCUGCUUCCCAACCCACAGAUGAAUU ..-......((----(((..(((....((((((((((((.(((....))))))))......-----------.----.............)))))))..))).)))))......... ( -30.95) >DroEre_CAF1 42959 96 + 1 GC-AGUGAAGU----GGGAGUGGCAGUAGCUGGAGUGGGCGUUACAGGACCCCACUAAGAA----------------AAAAACAGAGGCUUCCUGCUUCCCAACCCACAGAUGAAUU ..-.(((...(----((((..(((((.((((..((((((.(((....))))))))).....----------------.........))))..))))))))))...)))......... ( -32.49) >DroYak_CAF1 33192 112 + 1 GC-AGCGAAGU----GGGAGUGGCAGUAGCAGGAGUGGGCGUUACAGGACCCCACAUAAAAACACAAAAAAAUAAAAUAAAACAGAGGCUUCCUGCUUCCCAACCCACAGAUGAAUU ..-......((----(((..(((....((((((((((((.(((....))))))))...........................(....)..)))))))..))).)))))......... ( -31.60) >DroAna_CAF1 41990 90 + 1 GCCAGUGGGAGGCCGAGGAGUGGCAGUAGCAGGAGUGGGCGUCGGAGGACC---------------------------AAGAGAGAAGCUUCCUGCUUCCCAGCCCACGGACGAAUU .((.((.....((((.....))))....))....(((((((((....))).---------------------------....(.(((((.....))))).).))))))))....... ( -29.80) >consensus GC_AGUGAAGU____GGGAGUGGCAGUAGCAGGAGUGGGCGUUACAGGACCCCACCAAAAA___________U____AAAAACAGAGGCUUCCUGCUUCCCAACCCACAGAUGAAUU ...............(((..(((....((((((((((((.(((....))))))))............................((...)))))))))..))).)))........... (-20.81 = -21.83 + 1.03)
| Location | 14,425,381 – 14,425,477 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.85 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -19.79 |
| Energy contribution | -19.60 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.556050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
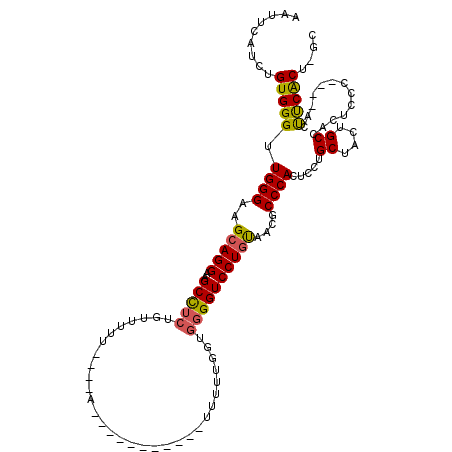
>3L_DroMel_CAF1 14425381 96 - 23771897 AAUUCAUCUGUGGGUUGGGAAGCAGGAAGCCUCU-UUUUU----A-----------UUUUUGGUGGGGUCCUGUAACGCCCACUGCUGCUACUGCCACUCCC----ACUUCACU-GA ......((.(((((.(((((.((((..(((....-.....----.-----------.....((((((((......)).)))))))))....))))...))))----).))))).-)) ( -29.36) >DroSec_CAF1 27738 97 - 1 AAUUCAUCUGUGGGUUGGGAAGCAGGAAGCCUCUUUUUUU----A-----------UUUUUGGUGGGGUCCUGUAACGCCCACUCCUGCUACUGCCACUCCC----CCUUCACU-GC .........(((((..(((((((((((.(((.........----.-----------.....))).((((........))))..)))))))........))))----..))))).-.. ( -26.86) >DroSim_CAF1 42542 97 - 1 AAUUCAUCUGUGGGUUGGGAAGCAGGAAGCCUCUGUUUUU----A-----------UUUUUGGUGGGGUCCUGUAACGCCCACUCCUGCUACUGCCACUCCC----ACUUCACU-GC .........(((((.(((.(((((((((((....))))..----.-----------.....((((((((......)).))))))))))))..).)))..)))----))......-.. ( -30.70) >DroEre_CAF1 42959 96 - 1 AAUUCAUCUGUGGGUUGGGAAGCAGGAAGCCUCUGUUUUU----------------UUCUUAGUGGGGUCCUGUAACGCCCACUCCAGCUACUGCCACUCCC----ACUUCACU-GC .........(((((.((((..((((((..((.(((.....----------------....))).))..))))))....)))).....((....))....)))----))......-.. ( -28.60) >DroYak_CAF1 33192 112 - 1 AAUUCAUCUGUGGGUUGGGAAGCAGGAAGCCUCUGUUUUAUUUUAUUUUUUUGUGUUUUUAUGUGGGGUCCUGUAACGCCCACUCCUGCUACUGCCACUCCC----ACUUCGCU-GC .........(((((.(((.((((((((.........................(((....)))(((((((......)).))))))))))))..).)))..)))----))......-.. ( -30.70) >DroAna_CAF1 41990 90 - 1 AAUUCGUCCGUGGGCUGGGAAGCAGGAAGCUUCUCUCUU---------------------------GGUCCUCCGACGCCCACUCCUGCUACUGCCACUCCUCGGCCUCCCACUGGC .......((((((((.(((((((.....)))))))..((---------------------------((....)))).))))))..........(((.......)))........)). ( -28.20) >consensus AAUUCAUCUGUGGGUUGGGAAGCAGGAAGCCUCUGUUUUU____A___________UUUUUGGUGGGGUCCUGUAACGCCCACUCCUGCUACUGCCACUCCC____ACUUCACU_GC .........(((((.((((..(((((..(((((...............................))))))))))....)))).....((....)).............))))).... (-19.79 = -19.60 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:02 2006