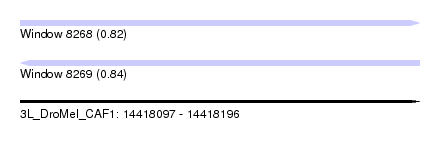
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,418,097 – 14,418,196 |
| Length | 99 |
| Max. P | 0.842992 |

| Location | 14,418,097 – 14,418,196 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
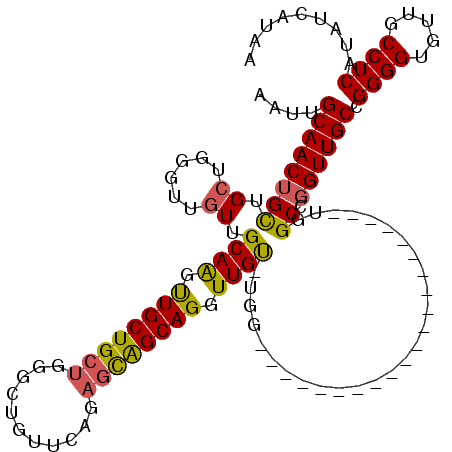
| Reading direction | forward |
| Mean pairwise identity | 80.63 |
| Mean single sequence MFE | -24.85 |
| Consensus MFE | -16.20 |
| Energy contribution | -17.53 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824150 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14418097 99 + 23771897 UUAUGAUAUGAGGCAACACCCCGGCAACCGGCCA---------------------CCAAACAACCUGCUGCUCUGAACAGCCCAGCAGCAGCUUGCAACAACCCAUCAGCAGUUGCAAUU ............(((((.....(((.....))).---------------------.....(((.((((((((((....))...)))))))).)))................))))).... ( -26.20) >DroSec_CAF1 20478 98 + 1 UUAUGAUAUGAGGCAACACCCCGGCAACCGGCCA---------------------CCA-ACAACCUGCUGCUCUGAACAGCCCAGCAGCAACUUGCAACAACCCAGCAGCAGUUGCAAUU ............(((((.....(((.....))).---------------------...-......((((((((((.......)))..((.....))........)))))))))))).... ( -24.80) >DroSim_CAF1 34667 98 + 1 UUAUGAUAUGAGGCAACACCCCGGCAACCGGCCA---------------------CCA-ACAACCUGCUGCUCCGAACAGACCAGCAGCAACUUGCAACAACCCAGCAGCAGUUGCAAUU ............(((((.....(((.....))).---------------------...-......(((((((.(.....)....((((....))))........)))))))))))).... ( -23.60) >DroEre_CAF1 39822 119 + 1 UUAUGGUAUGAGGCAACACCCCGGCAACAGGCAACGGGCAACAGGCAACCGACCACCA-ACAACCUGCUGCUUGAAACAGCUCAGCAGCAACUUGCAACAACCCAGCAGCAGUUGCAAUU ...((((.....((.....((((....).(....))))......)).....))))...-......(((((((.((......)))))))))..(((((((............))))))).. ( -32.80) >DroYak_CAF1 25929 98 + 1 UUAUGAUAUGAGGCAACACCCCGGCAACCGGCAA---------------------CCA-ACAACCUGCUGCUUGAAGCAGCCCAGCAGCAAGUUGCAACAACCCAGCAGCAGUUGCAAUU ............(((((...((((...))))...---------------------...-......(((((((((..(((((..........)))))..))....)))))))))))).... ( -27.30) >DroAna_CAF1 35783 81 + 1 UUAUGAUAUGAGGCAACACCCCAGCAACCAGCAA---------------------CCA-GCAACCUGCCACC-------------CAGCAACCUGCAAC----CUGCAACAGUUGCAAUU ...........(....)......(((((..(((.---------------------...-(((...(((....-------------..)))...)))...----.)))....))))).... ( -14.40) >consensus UUAUGAUAUGAGGCAACACCCCGGCAACCGGCAA_____________________CCA_ACAACCUGCUGCUCGGAACAGCCCAGCAGCAACUUGCAACAACCCAGCAGCAGUUGCAAUU ............(((((...((((...)))).............................(((..(((((((...........)))))))..)))................))))).... (-16.20 = -17.53 + 1.33)

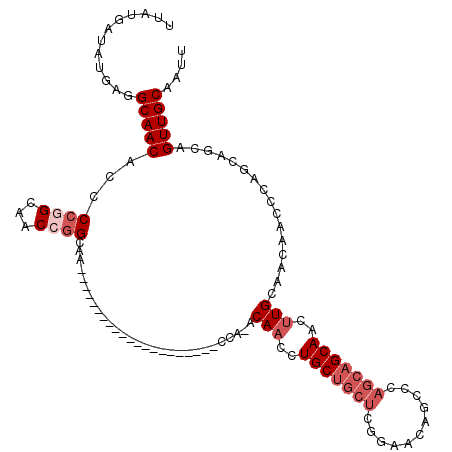
| Location | 14,418,097 – 14,418,196 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.63 |
| Mean single sequence MFE | -35.75 |
| Consensus MFE | -26.25 |
| Energy contribution | -26.08 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14418097 99 - 23771897 AAUUGCAACUGCUGAUGGGUUGUUGCAAGCUGCUGCUGGGCUGUUCAGAGCAGCAGGUUGUUUGG---------------------UGGCCGGUUGCCGGGGUGUUGCCUCAUAUCAUAA ....((((((.(....)(((..((((((.((((((((.((....))..)))))))).))))..))---------------------..))))))))).((((.....))))......... ( -35.80) >DroSec_CAF1 20478 98 - 1 AAUUGCAACUGCUGCUGGGUUGUUGCAAGUUGCUGCUGGGCUGUUCAGAGCAGCAGGUUGU-UGG---------------------UGGCCGGUUGCCGGGGUGUUGCCUCAUAUCAUAA ....((((((((..(((.......((((.((((((((.((....))..)))))))).))))-)))---------------------..).))))))).((((.....))))......... ( -34.81) >DroSim_CAF1 34667 98 - 1 AAUUGCAACUGCUGCUGGGUUGUUGCAAGUUGCUGCUGGUCUGUUCGGAGCAGCAGGUUGU-UGG---------------------UGGCCGGUUGCCGGGGUGUUGCCUCAUAUCAUAA ....((((((((..(..(((....))......((((((.(((....))).))))))....)-..)---------------------..).))))))).((((.....))))......... ( -35.60) >DroEre_CAF1 39822 119 - 1 AAUUGCAACUGCUGCUGGGUUGUUGCAAGUUGCUGCUGAGCUGUUUCAAGCAGCAGGUUGU-UGGUGGUCGGUUGCCUGUUGCCCGUUGCCUGUUGCCGGGGUGUUGCCUCAUACCAUAA ....(((((.((.((.((((....((((.(((((((((((....))).)))))))).))))-.((..(....)..))....)))))).))..))))).((((.....))))......... ( -41.90) >DroYak_CAF1 25929 98 - 1 AAUUGCAACUGCUGCUGGGUUGUUGCAACUUGCUGCUGGGCUGCUUCAAGCAGCAGGUUGU-UGG---------------------UUGCCGGUUGCCGGGGUGUUGCCUCAUAUCAUAA ....((((((((.((((.......(((((((((((((.((.....)).)))))))))))))-)))---------------------).).))))))).((((.....))))......... ( -36.21) >DroAna_CAF1 35783 81 - 1 AAUUGCAACUGUUGCAG----GUUGCAGGUUGCUG-------------GGUGGCAGGUUGC-UGG---------------------UUGCUGGUUGCUGGGGUGUUGCCUCAUAUCAUAA ....((((((...((((----.(.((((.(((((.-------------...))))).))))-.).---------------------)))).))))))(((((.....)))))........ ( -30.20) >consensus AAUUGCAACUGCUGCUGGGUUGUUGCAAGUUGCUGCUGGGCUGUUCAGAGCAGCAGGUUGU_UGG_____________________UGGCCGGUUGCCGGGGUGUUGCCUCAUAUCAUAA ....((((((((.((......)).((((.((((((((...........)))))))).))))...........................)).)))))).((((.....))))......... (-26.25 = -26.08 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:59 2006