
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,384,034 – 14,384,162 |
| Length | 128 |
| Max. P | 0.975977 |

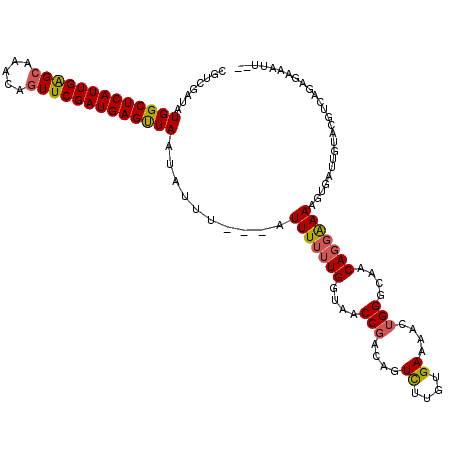
| Location | 14,384,034 – 14,384,150 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.34 |
| Mean single sequence MFE | -30.85 |
| Consensus MFE | -20.34 |
| Energy contribution | -20.65 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14384034 116 - 23771897 CGUCGAUAUGGCUCAUUGAGCAAACAGUUCGAUGAGUUAAUAUUUAAUAUUUUUUGGUAACCGACAGUCUUGUGAAAACUGGGCAACAGGAAAAGUGAUUGUACCUUAGAGUAAUU-- ....(((((((((((((((((.....)))))))))))).)))))...((((((..((((.((....((((.((....)).))))....))..((....)).))))..))))))...-- ( -31.50) >DroSim_CAF1 5360 112 - 1 CGUCGAUAUGGCUCAUUGGGCAAACAGUUCGAUGAGUUAAUAUUU---AUUUUUUGGUAACCGACAGUCUUGUGAAAAGUGGGCAACAGGAAAAGUGAUUGUACUUC-AAGUAAUU-- ....((..(((((((((((((.....)))))))))))))(((.((---(((((((.((..((.((..((....))...)).))..))..))))))))).)))...))-........-- ( -27.60) >DroEre_CAF1 5540 112 - 1 CGUCG-UGUGGCUCAUUGGGCAAACAGUUCGAUGAGUUAAUAUUU---AUUUUUUGGCAACCGGCAGUCUUGUGAAAACUGGGCAACAGGAAAAGUUAAAGUACGUCAGAGAAAUU-- .....-..(((((((((((((.....)))))))))))))......---.(((((((((..((....((((.((....)).))))....))....((......)))))))))))...-- ( -31.80) >DroAna_CAF1 4957 107 - 1 CGCCGAUAUGGCUCAUUGAGCAAAUACUUCGAUGAGCUAA--GUC---AUUUUUUGAAAA--GUCAGUUGUCUGAAAGC----CAGCACUGAAGGAAGUUGUCUGUUCGAGAAAUAAA ....(((.((((((((((((.......)))))))))))).--)))---((((((((((..--.(((((.(.(((.....----))))))))).(((.....))).))))))))))... ( -32.50) >consensus CGUCGAUAUGGCUCAUUGAGCAAACAGUUCGAUGAGUUAAUAUUU___AUUUUUUGGUAACCGACAGUCUUGUGAAAACUGGGCAACAGGAAAAGUGAUUGUACGUCAGAGAAAUU__ ........(((((((((((((.....)))))))))))))..........(((((((....(((....((....))....)))....)))))))......................... (-20.34 = -20.65 + 0.31)


| Location | 14,384,072 – 14,384,162 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 75.63 |
| Mean single sequence MFE | -24.91 |
| Consensus MFE | -17.30 |
| Energy contribution | -17.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.920575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14384072 90 - 23771897 ---UGGGGGAUA-----------------UGGCGUCGAUAUGGCUCAUUGAGCAAACAGUUCGAUGAGUUAAUAUUUAAUAUUUUUUGGUAACCGACAGUCUUGUGAAAA ---......(((-----------------.(((((((...(((((((((((((.....)))))))))))))........((((....))))..)))).))).)))..... ( -23.70) >DroSim_CAF1 5397 87 - 1 ---UGGGGGAUA-----------------UGGCGUCGAUAUGGCUCAUUGGGCAAACAGUUCGAUGAGUUAAUAUUU---AUUUUUUGGUAACCGACAGUCUUGUGAAAA ---......(((-----------------.(((((((...(((((((((((((.....)))))))))))))....((---(((....))))).)))).))).)))..... ( -23.20) >DroEre_CAF1 5578 86 - 1 ---UGGGGGAUA-----------------CGGCGUCG-UGUGGCUCAUUGGGCAAACAGUUCGAUGAGUUAAUAUUU---AUUUUUUGGCAACCGGCAGUCUUGUGAAAA ---.(.(((((.-----------------(((.((((-..(((((((((((((.....)))))))))))))......---......))))..)))...))))).)..... ( -23.04) >DroAna_CAF1 4993 103 - 1 GACUGAGGAAUACUAUAUGAUGUAGUAACCGGCGCCGAUAUGGCUCAUUGAGCAAAUACUUCGAUGAGCUAA--GUC---AUUUUUUGAAAA--GUCAGUUGUCUGAAAG ((((((((..((((((.....)))))).))......(((.((((((((((((.......)))))))))))).--)))---............--.))))))......... ( -29.70) >consensus ___UGGGGGAUA_________________CGGCGUCGAUAUGGCUCAUUGAGCAAACAGUUCGAUGAGUUAAUAUUU___AUUUUUUGGUAACCGACAGUCUUGUGAAAA ..............................((((((....(((((((((((((.....)))))))))))))................(.....)))).)))......... (-17.30 = -17.30 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:47 2006