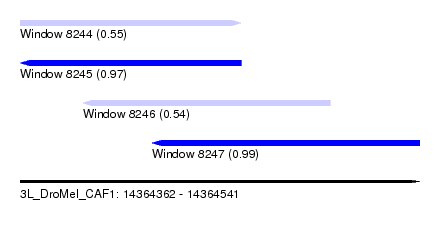
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,364,362 – 14,364,541 |
| Length | 179 |
| Max. P | 0.989210 |

| Location | 14,364,362 – 14,364,461 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 86.52 |
| Mean single sequence MFE | -17.32 |
| Consensus MFE | -13.58 |
| Energy contribution | -13.74 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14364362 99 + 23771897 UCCAUCUGCAGGCCUACCCACAAUAGUUAACAGCAUUGUUACACUUUUAAACGGUAUACUUACACCCAUUCCUAUUGAUUCGAAUGGGUAUAUUGCCAU ..........(((.((((......(((((((......)))).))).......)))).......((((((((..........)))))))).....))).. ( -20.92) >DroSec_CAF1 188596 99 + 1 UCCAUUUGCAGGCCUACCCACAAUAGUUAACAGCAUUGUUACACUUUUAAACGGUAUACUUACACCCAUUCCAAGUGAUUCGAAUGGGUAUAUUAACAU .......(.(((..((((......(((((((......)))).))).......))))..))).)((((((((.((....)).)))))))).......... ( -20.02) >DroSim_CAF1 184273 99 + 1 UCCAUCUGCAGGCCUACCCACAAUAGUUAACAGCAUUGUUACACUUUUAAACGGUAUACUUACACCCAUUCCAAGUGAUUCGAAUGGGUAUAUUAACAU .......(.(((..((((......(((((((......)))).))).......))))..))).)((((((((.((....)).)))))))).......... ( -20.02) >DroEre_CAF1 187282 88 + 1 UC-----------CUACCCACAAUAGUUAACAGCAUUGUUACACUUUUAAACGGUAUACUUACACCCAGUCCCAGUGAUUCGACAGGGUAUACUGCCAU ..-----------.((((((((((.((.....)))))))..((((.......(((........))).......))))........)))))......... ( -14.24) >DroYak_CAF1 194882 88 + 1 UU-----------CUAUCCACAAUAGUUAACAGCAUUGUUACACCUUUAAACGGUAUACUUACACCCAGUCCUAGUGAUUCGACAGGGCAUACUGACAU ..-----------......(((((.((.....)))))))............((((((.......(((.(((..(....)..))).))).)))))).... ( -11.40) >consensus UCCAU_UGCAGGCCUACCCACAAUAGUUAACAGCAUUGUUACACUUUUAAACGGUAUACUUACACCCAUUCCAAGUGAUUCGAAUGGGUAUAUUGACAU ..............((((......(((((((......)))).))).......)))).......((((((((..........)))))))).......... (-13.58 = -13.74 + 0.16)

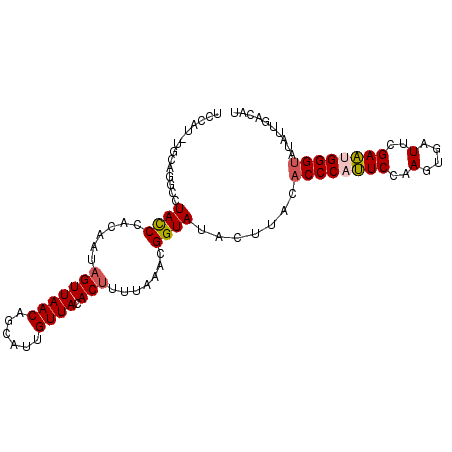
| Location | 14,364,362 – 14,364,461 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 86.52 |
| Mean single sequence MFE | -26.76 |
| Consensus MFE | -20.90 |
| Energy contribution | -20.70 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14364362 99 - 23771897 AUGGCAAUAUACCCAUUCGAAUCAAUAGGAAUGGGUGUAAGUAUACCGUUUAAAAGUGUAACAAUGCUGUUAACUAUUGUGGGUAGGCCUGCAGAUGGA .......(((((((((((..........)))))))))))......((((((...(((.(((((....))))))))...(..((....))..))))))). ( -29.50) >DroSec_CAF1 188596 99 - 1 AUGUUAAUAUACCCAUUCGAAUCACUUGGAAUGGGUGUAAGUAUACCGUUUAAAAGUGUAACAAUGCUGUUAACUAUUGUGGGUAGGCCUGCAAAUGGA .......(((((((((((.((....)).)))))))))))......((((((...(((.(((((....))))))))...(..((....))..))))))). ( -30.90) >DroSim_CAF1 184273 99 - 1 AUGUUAAUAUACCCAUUCGAAUCACUUGGAAUGGGUGUAAGUAUACCGUUUAAAAGUGUAACAAUGCUGUUAACUAUUGUGGGUAGGCCUGCAGAUGGA .......(((((((((((.((....)).)))))))))))......((((((...(((.(((((....))))))))...(..((....))..))))))). ( -31.00) >DroEre_CAF1 187282 88 - 1 AUGGCAGUAUACCCUGUCGAAUCACUGGGACUGGGUGUAAGUAUACCGUUUAAAAGUGUAACAAUGCUGUUAACUAUUGUGGGUAG-----------GA ...((..(((((((.(((..........))).))))))).)).((((...(((.(((.(((((....)))))))).)))..)))).-----------.. ( -20.60) >DroYak_CAF1 194882 88 - 1 AUGUCAGUAUGCCCUGUCGAAUCACUAGGACUGGGUGUAAGUAUACCGUUUAAAGGUGUAACAAUGCUGUUAACUAUUGUGGAUAG-----------AA .((((..(((((((.(((..........))).)))))))..((((((.......))))))((((((........)))))).)))).-----------.. ( -21.80) >consensus AUGUCAAUAUACCCAUUCGAAUCACUAGGAAUGGGUGUAAGUAUACCGUUUAAAAGUGUAACAAUGCUGUUAACUAUUGUGGGUAGGCCUGCA_AUGGA .......(((((((((((..........)))))))))))....((((...(((.(((.(((((....)))))))).)))..)))).............. (-20.90 = -20.70 + -0.20)


| Location | 14,364,390 – 14,364,501 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.91 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -16.00 |
| Energy contribution | -15.96 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14364390 111 - 23771897 GAAAGAUAUUCUGAGCUCUGAUUGAGUAUAAUUCUGAAAAAUGGCAAUAUACCCAUUCGAAUCAAUAGGAAUGGGUGUAAGUAUACCGUUUAAAAGUGUAACAAUGCUGUU (((..((((((............))))))..))).....(((((((.(((((((((((..........)))))))))))..(((((.........)))))....))))))) ( -26.00) >DroSec_CAF1 188624 95 - 1 GAAAGAUGUUCU----------------UAAUUUUGACAUAUGUUAAUAUACCCAUUCGAAUCACUUGGAAUGGGUGUAAGUAUACCGUUUAAAAGUGUAACAAUGCUGUU ...(((((....----------------.....(((((....)))))(((((((((((.((....)).))))))))))).......)))))...(((((....)))))... ( -20.70) >DroSim_CAF1 184301 95 - 1 GAAAGAUGUUCU----------------UAAUUUUGACAUAUGUUAAUAUACCCAUUCGAAUCACUUGGAAUGGGUGUAAGUAUACCGUUUAAAAGUGUAACAAUGCUGUU ...(((((....----------------.....(((((....)))))(((((((((((.((....)).))))))))))).......)))))...(((((....)))))... ( -20.70) >DroEre_CAF1 187299 109 - 1 UAAAAGUAUUCUGAGCUCUGAUAGA-UACUA-UCUGCUAAAUGGCAGUAUACCCUGUCGAAUCACUGGGACUGGGUGUAAGUAUACCGUUUAAAAGUGUAACAAUGCUGUU ....((((((.(..(((..((..((-((((.-.(((((....)))))(((((((.(((..........))).)))))))))))).)..))....)))...).))))))... ( -26.40) >DroYak_CAF1 194899 106 - 1 ----AGGAUUCUCAGCUCAGAUAGAGUAUUA-UCUGCAAAAUGUCAGUAUGCCCUGUCGAAUCACUAGGACUGGGUGUAAGUAUACCGUUUAAAGGUGUAACAAUGCUGUU ----........((((.(((((((....)))-)))).....(((...(((((((.(((..........))).)))))))..((((((.......)))))))))..)))).. ( -26.70) >consensus GAAAGAUAUUCU_AGCUC_GAU_GA_UAUAAUUCUGAAAAAUGUCAAUAUACCCAUUCGAAUCACUAGGAAUGGGUGUAAGUAUACCGUUUAAAAGUGUAACAAUGCUGUU .................................(((((....)))))(((((((((((..........)))))))))))..(((((.........)))))........... (-16.00 = -15.96 + -0.04)

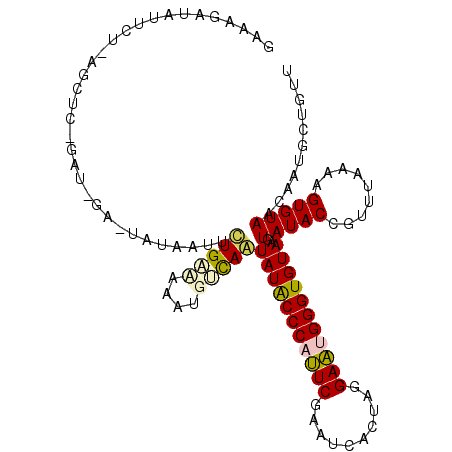
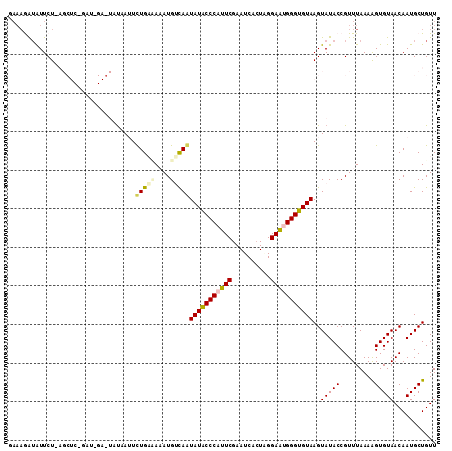
| Location | 14,364,421 – 14,364,541 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.70 |
| Mean single sequence MFE | -28.63 |
| Consensus MFE | -15.36 |
| Energy contribution | -14.92 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14364421 120 - 23771897 AUUUCGUUGCUUAAGAAAAGGAUUGGCUUGAAGAGUUUACGAAAGAUAUUCUGAGCUCUGAUUGAGUAUAAUUCUGAAAAAUGGCAAUAUACCCAUUCGAAUCAAUAGGAAUGGGUGUAA .....((((((.......((((((((((..(((((((((.(((.....)))))))))))..)..))).))))))).......))))))((((((((((..........)))))))))).. ( -37.74) >DroSec_CAF1 188655 104 - 1 AUUUCGUUGCUUAAGAAUAGGAUUGGCUUGAAAAGUUUACGAAAGAUGUUCU----------------UAAUUUUGACAUAUGUUAAUAUACCCAUUCGAAUCACUUGGAAUGGGUGUAA ..........(((((((((.....((((.....))))..(....).))))))----------------)))..(((((....)))))(((((((((((.((....)).))))))))))). ( -26.00) >DroSim_CAF1 184332 104 - 1 AUUUCGUUGCUUAAGAAUAGGAUUGGCUUGAAAAGUUUACGAAAGAUGUUCU----------------UAAUUUUGACAUAUGUUAAUAUACCCAUUCGAAUCACUUGGAAUGGGUGUAA ..........(((((((((.....((((.....))))..(....).))))))----------------)))..(((((....)))))(((((((((((.((....)).))))))))))). ( -26.00) >DroEre_CAF1 187330 118 - 1 UUUUUCUUGCUCAAGCAUGGGAAUGGCUUAAGGAGUUUACUAAAAGUAUUCUGAGCUCUGAUAGA-UACUA-UCUGCUAAAUGGCAGUAUACCCUGUCGAAUCACUGGGACUGGGUGUAA .((..(.(((....))).)..))...((...((((((((............))))))))...)).-.....-.(((((....)))))(((((((.(((..........))).))))))). ( -30.10) >DroYak_CAF1 194930 105 - 1 AUUUACCUGCUUAAGAAUAGGAAAGGUUAU--------------AGGAUUCUCAGCUCAGAUAGAGUAUUA-UCUGCAAAAUGUCAGUAUGCCCUGUCGAAUCACUAGGACUGGGUGUAA ...(((((.((((....))))...((((.(--------------(((((((........(((((.((((..-.((((.....).))).)))).)))))))))).))).)))))))))... ( -23.30) >consensus AUUUCGUUGCUUAAGAAUAGGAUUGGCUUGAAAAGUUUACGAAAGAUAUUCU_AGCUC_GAU_GA_UAUAAUUCUGAAAAAUGUCAAUAUACCCAUUCGAAUCACUAGGAAUGGGUGUAA .((((...(((.............)))..))))........................................(((((....)))))(((((((((((..........))))))))))). (-15.36 = -14.92 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:38 2006