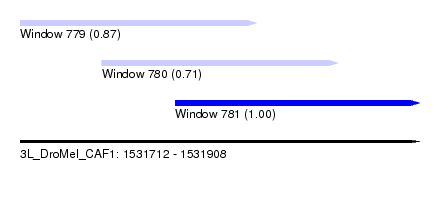
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,531,712 – 1,531,908 |
| Length | 196 |
| Max. P | 0.998905 |

| Location | 1,531,712 – 1,531,828 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.63 |
| Mean single sequence MFE | -37.98 |
| Consensus MFE | -30.08 |
| Energy contribution | -28.92 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1531712 116 + 23771897 AAGAUCAGGCGGUGUGAUUGUCAGACAUGCAGGGUGUUUGUCGAGUGCUAACCAUAAUUAAAGGCGAAUUU----GUCUUAUCGCUGGAAACAGUGAAACAUGGCUCGUUUCCGCCAAAC .......(((((..((((((.(((((((.....))))))).)))..((((..........((((((....)----))))).((((((....))))))....)))))))...))))).... ( -38.00) >DroSec_CAF1 10521 116 + 1 AAGAUCGGGCGGUGCGUUUAUCAGACAUGCAGGGUGUUUGCCGAGUGCUAACCACAAUUAAAGGCGAAUUG----GUCUUAUCGCUGGAAACAGUGAAACAUGGCUCGUUUCCGCCAAAC .......(((((.(((......((((((.....))))))((((.((..............(((((......----))))).((((((....)))))).)).)))).)))..))))).... ( -36.80) >DroSim_CAF1 10648 116 + 1 AAGAUCGGGCGGUGCGAUUAUCAGACAUGCAGGGUGUUUGCCGAGUGCUAACCAUAAUUAAAGGCGAAUUG----GUCUUAUCGCUGGAAACAGUGAAACACGGCUCGUUUCCGCCAAAC .......(((((.((((.....((((((.....))))))((((.((..............(((((......----))))).((((((....)))))).)).))))))))..))))).... ( -40.40) >DroEre_CAF1 10536 116 + 1 AAGAUCAGGCGGUGCGAUUAUCGCGCUUGCUGGGUGUUUGCUGAGUGCUAACCAUAAUUAAAGGUAAAUGU----GUCUUAUCGCUGGAAACAGUGAAACACGGCUCGUUUCCGCCAAAC .......(((((.((((.....((((((((.........)).))))))..(((.........)))...(((----((....((((((....)))))).)))))..))))..))))).... ( -38.80) >DroYak_CAF1 10730 120 + 1 AAGAUCGGGCGGUGUGAUUAUCGGGCUUGCAGCGUGUUUGCUAAGUGCUAACCAUAAUUAAAGGUGAAUUUACCAGUCUUAUCGCUGGUAACAGUGAAACAUGGCCCGUUUCCGCCAAGC ..((.(((((.((((((((...(.((((((((.....)))).)))).)..............((((....))))))))...((((((....)))))).)))).)))))..))........ ( -35.90) >consensus AAGAUCGGGCGGUGCGAUUAUCAGACAUGCAGGGUGUUUGCCGAGUGCUAACCAUAAUUAAAGGCGAAUUU____GUCUUAUCGCUGGAAACAGUGAAACAUGGCUCGUUUCCGCCAAAC .......(((((.(((......((((((.....))))))((((.((..............(((((..........))))).((((((....)))))).)).)))).)))..))))).... (-30.08 = -28.92 + -1.16)

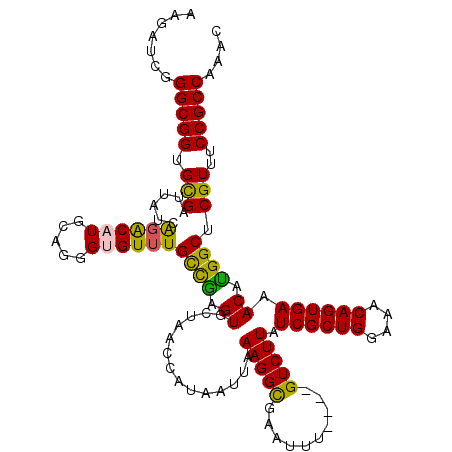
| Location | 1,531,752 – 1,531,868 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.03 |
| Mean single sequence MFE | -32.68 |
| Consensus MFE | -27.28 |
| Energy contribution | -27.08 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1531752 116 + 23771897 UCGAGUGCUAACCAUAAUUAAAGGCGAAUUU----GUCUUAUCGCUGGAAACAGUGAAACAUGGCUCGUUUCCGCCAAACGCAUUUAUCGUGUUUCCAGAUACCCGCGAUUCGCGAUUCG ..((((((............((((((....)----))))).((((((....))))))....((((........))))...))))))((((((..(((........).))..))))))... ( -32.90) >DroSec_CAF1 10561 116 + 1 CCGAGUGCUAACCACAAUUAAAGGCGAAUUG----GUCUUAUCGCUGGAAACAGUGAAACAUGGCUCGUUUCCGCCAAACGCAUUUAUCGUGUUUCCAGAUACCCGCGAUUCGCGAUUCG ..((((((............(((((......----))))).((((((....))))))....((((........))))...))))))((((((..(((........).))..))))))... ( -33.00) >DroSim_CAF1 10688 116 + 1 CCGAGUGCUAACCAUAAUUAAAGGCGAAUUG----GUCUUAUCGCUGGAAACAGUGAAACACGGCUCGUUUCCGCCAAACGCAUUUAUCGUGUUUCCAGAUACCCGCGAUUCGCGAUUCG .(((((.((............))((((((((----....((((((((....))))(((((((((..(((((.....)))))......)))))))))..))))....)))))))).))))) ( -32.50) >DroEre_CAF1 10576 116 + 1 CUGAGUGCUAACCAUAAUUAAAGGUAAAUGU----GUCUUAUCGCUGGAAACAGUGAAACACGGCUCGUUUCCGCCAAACGCAUUUAUCGUGUUUCCAGAUACUCGCGAUUCGCGUUUCG ..((((((..............(((((((((----((....((((((....)))))).....(((........)))..)))))))))))(((((....)))))..)).))))........ ( -35.00) >DroYak_CAF1 10770 120 + 1 CUAAGUGCUAACCAUAAUUAAAGGUGAAUUUACCAGUCUUAUCGCUGGUAACAGUGAAACAUGGCCCGUUUCCGCCAAGCGCAUUUAUCGUGUUUCCAGAUACUCGCGAUUCGCGAUUCG ....(((((.............((((....)))).......((((((....))))))....((((........))))))))).......(((((....)))))(((((...))))).... ( -30.00) >consensus CCGAGUGCUAACCAUAAUUAAAGGCGAAUUU____GUCUUAUCGCUGGAAACAGUGAAACAUGGCUCGUUUCCGCCAAACGCAUUUAUCGUGUUUCCAGAUACCCGCGAUUCGCGAUUCG ..((((((............(((((..........))))).((((((....)))))).....(((........)))....))))))((((((..(((........).))..))))))... (-27.28 = -27.08 + -0.20)

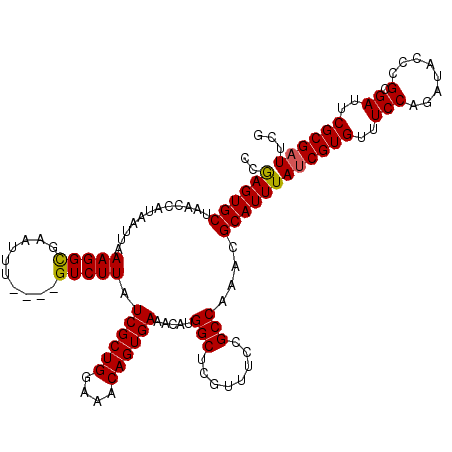
| Location | 1,531,788 – 1,531,908 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -41.76 |
| Consensus MFE | -38.08 |
| Energy contribution | -38.96 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.28 |
| SVM RNA-class probability | 0.998905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
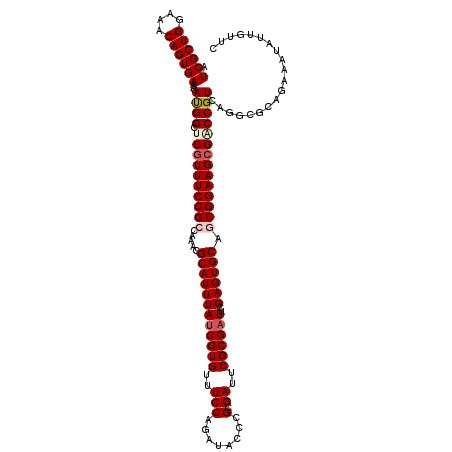
>3L_DroMel_CAF1 1531788 120 + 23771897 AUCGCUGGAAACAGUGAAACAUGGCUCGUUUCCGCCAAACGCAUUUAUCGUGUUUCCAGAUACCCGCGAUUCGCGAUUCGAGUGCAGCGGAAGCGGCCGUCAGGCGCAGAAAUAUUGUUC .((((((....))))))...(((((.(((((((((.....((((((((((((..(((........).))..))))))..)))))).)))))))))))))).................... ( -46.30) >DroSec_CAF1 10597 120 + 1 AUCGCUGGAAACAGUGAAACAUGGCUCGUUUCCGCCAAACGCAUUUAUCGUGUUUCCAGAUACCCGCGAUUCGCGAUUCGAGUGCAGCGGAAGCGACCGUCAGGCGCAGAAAUAUUGUUC .((((((....))))))...((((.((((((((((.....((((((((((((..(((........).))..))))))..)))))).)))))))))))))).................... ( -44.20) >DroSim_CAF1 10724 120 + 1 AUCGCUGGAAACAGUGAAACACGGCUCGUUUCCGCCAAACGCAUUUAUCGUGUUUCCAGAUACCCGCGAUUCGCGAUUCGAGUGCAGCGGAAGCGACCGUCAGGCGCAGAAAUAUUGUUC .((((((....))))))...((((.((((((((((.....((((((((((((..(((........).))..))))))..)))))).)))))))))))))).................... ( -46.00) >DroEre_CAF1 10612 120 + 1 AUCGCUGGAAACAGUGAAACACGGCUCGUUUCCGCCAAACGCAUUUAUCGUGUUUCCAGAUACUCGCGAUUCGCGUUUCGAGUGCAACGGAAGAGCCCGUCACCCACAUAAAUAUGGUUC .((((((....)))))).....(((((...((((..((((((.......))))))...(.((((((.(((....))).)))))))..)))).)))))......(((........)))... ( -36.30) >DroYak_CAF1 10810 120 + 1 AUCGCUGGUAACAGUGAAACAUGGCCCGUUUCCGCCAAGCGCAUUUAUCGUGUUUCCAGAUACUCGCGAUUCGCGAUUCGAGUGCAACGGAAGCGCCCGUCAGCCGCAGAAAUAUUGUUC .((((((....))))))....((((.((((((((....((((....((((((..(((........).))..))))))....))))..))))))))...)))).................. ( -36.00) >consensus AUCGCUGGAAACAGUGAAACAUGGCUCGUUUCCGCCAAACGCAUUUAUCGUGUUUCCAGAUACCCGCGAUUCGCGAUUCGAGUGCAGCGGAAGCGACCGUCAGGCGCAGAAAUAUUGUUC .((((((....))))))...((((.((((((((((.....((((((((((((..(((........).))..))))))..)))))).)))))))))))))).................... (-38.08 = -38.96 + 0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:09 2006