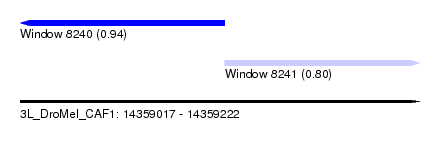
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,359,017 – 14,359,222 |
| Length | 205 |
| Max. P | 0.938625 |

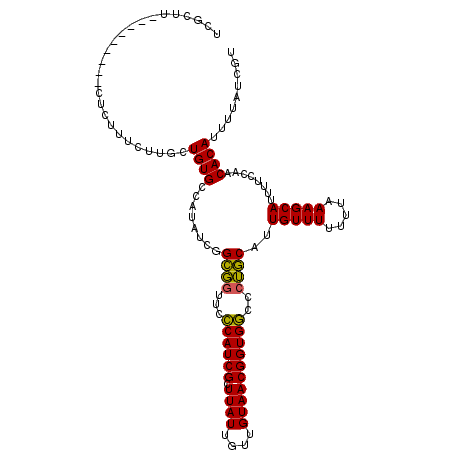
| Location | 14,359,017 – 14,359,122 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 83.48 |
| Mean single sequence MFE | -18.91 |
| Consensus MFE | -18.93 |
| Energy contribution | -17.93 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.13 |
| Mean z-score | -0.49 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14359017 105 - 23771897 UCGCUU---------CUCUUUCUUGCUGUGCCAUAUCGGCGGUUCCCAUCGCUUAUUGUUGUAACGGUGGCCCUGCAUUGUUUUUUUAAAGCAUUUUCCAACACAUUUUAUCGU ..((((---------........(((...(((.....)))((...((((((.((((....)))))))))).)).)))...........))))...................... ( -19.41) >DroSec_CAF1 183521 105 - 1 UCGCUU---------CUCUUUCUUGCUGUGCCAUAUCGGCGGUUCCCAUCGCUUAUUGUUGUAACGGUGGCCCUGCAUUGUUUUUUUUAAGCAUUUUCCAACACAUUUUAUCGU ..((((---------........(((...(((.....)))((...((((((.((((....)))))))))).)).)))...........))))...................... ( -19.41) >DroPer_CAF1 195620 111 - 1 UUGCUUUUUUUCUCUCUCUAUCUUGCUGUGCGCUAUCGGGCAAAAGCAUCGCUUAUUGUUGUAACGGUGCC-CGCCAUUGUUU--UUCAAGCAUUUUCCAACACAUUUUAUCGU .(((((..................((.....)).....(((....((((((.((((....)))))))))).-.))).......--...)))))..................... ( -17.90) >consensus UCGCUU_________CUCUUUCUUGCUGUGCCAUAUCGGCGGUUCCCAUCGCUUAUUGUUGUAACGGUGGCCCUGCAUUGUUUUUUUAAAGCAUUUUCCAACACAUUUUAUCGU ..........................((((........((((...((((((.((((....))))))))))..))))..(((((.....)))))........))))......... (-18.93 = -17.93 + -1.00)


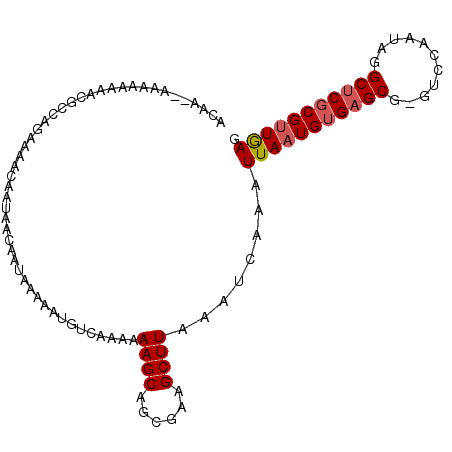
| Location | 14,359,122 – 14,359,222 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 87.68 |
| Mean single sequence MFE | -16.88 |
| Consensus MFE | -13.21 |
| Energy contribution | -13.78 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14359122 100 + 23771897 ACAA--AAAAAAACCGCCAGAAAACCAUAACAAUAAAAAUGUCAAAAAAGCAGCGAAGCUUAAAUCAAAUUAAUGUGAGCG-GUCCAAUAGGCUCGCGUUAAG ....--.........................................((((......))))........(((((((((((.-.........))))))))))). ( -16.40) >DroSec_CAF1 183626 99 + 1 ACAA--AA-AAAACCGCCAGAAAACCAUAACAAUAAAAAUGUCAAAAAAGCAGCGAAGCUUAAAUCAAAUUAAUGUGAGCG-GUCCAAUAGGCUCGCGUUGAG ....--..-......................................((((......))))........(((((((((((.-.........))))))))))). ( -16.50) >DroEre_CAF1 182030 102 + 1 GUUAAAAAAAAACAAGCCAGCAAACAAUAACAAUAAAAAUGUCAAAAAAGCAGCGAUGCUUAAAUCAAAUUAAUGUGAGCG-GUCCAAUAGGCUCGCGUUGAG ...............................................(((((....)))))........(((((((((((.-.........))))))))))). ( -18.00) >DroAna_CAF1 174811 97 + 1 ------AAAAACCACACCAGAAAACAAUAACAAUAAAAAUGUCAAAAAAGCAGCCGAGCUUAAAUCAAAUUAAUGUGAGCGGGUCCAAUAGGCGCUCGCUGAG ------......((((.............(((.......))).....((((......))))............))))(((((((((....)).)))))))... ( -16.60) >consensus ACAA__AAAAAAAACGCCAGAAAACAAUAACAAUAAAAAUGUCAAAAAAGCAGCGAAGCUUAAAUCAAAUUAAUGUGAGCG_GUCCAAUAGGCUCGCGUUGAG ...............................................((((......))))........(((((((((((...........))))))))))). (-13.21 = -13.78 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:32 2006