| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,358,094 – 14,358,212 |
| Length | 118 |
| Max. P | 0.528611 |

| Location | 14,358,094 – 14,358,212 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.64 |
| Mean single sequence MFE | -28.86 |
| Consensus MFE | -14.06 |
| Energy contribution | -13.73 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.49 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
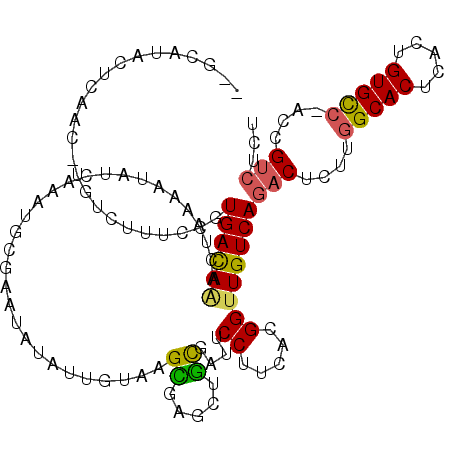
>3L_DroMel_CAF1 14358094 118 - 23771897 CAACAUACUCAAC-UGUCUAUCCGUGAUAAACUAAAAUAUAAAAUGCGAAUAUAUUUUAAGUGAGCUGCGAUCCCUUCACGGUUGUCAGACUCUUGGCACUCACUGUGCC-ACCGUCUCU ............(-((.(...((((((.....(((((((((.........))))))))).((.....)).......))))))..).))).....((((((.....)))))-)........ ( -26.00) >DroSec_CAF1 182629 115 - 1 ---CAUUCUCAAU-UGCCUUUCCGUGACAAACUAAAAUAUCAAAGGAGAAUAUAUUGUAAGUGAGCUACGAUCCCUUCACGGUUGUCAGACUCUUGGCACUCACUGUGCC-ACCGUCUCU ---((((..((((-....(((((.(((............)))..)))))....))))..))))....((((((.......)))))).((((...((((((.....)))))-)..)))).. ( -24.30) >DroEre_CAF1 181005 115 - 1 --GCAU-CUCAAC-UGUCUUUCCGUGACAAACUAAAAUAUCAAAUGCGAAUAUAUCGUAAGCGAGCUGCGAUUCCUUCACGGUUGUCAGACUCUUGGCACUCACUGUGCC-ACUGCCUCA --(((.-.....(-((.(...((((((.................(((((.....))))).((.....)).......))))))..).))).....((((((.....)))))-).))).... ( -29.60) >DroYak_CAF1 188525 116 - 1 --GCAUACUCAAC-UGUCUUUCCGUGACAUACUAAAAUAUCAAAUGCGAAUAUAUUGUAAGUGAGCUGCGAUUCCUUCACGGUUGUCAGACUCUUGGCACUCACUGUGCC-ACCGUCUCU --..........(-((.(...((((((............(((..(((((.....)))))..)))............))))))..).))).....((((((.....)))))-)........ ( -27.85) >DroAna_CAF1 173964 116 - 1 C-G-AAUCUGAAA-UGUCACUCCUUGACAAACUGAAACACUAAAUCAGAAUUUGUUUUAAGCAAACAGUUUGUCCCUUACGGUUGUCAGACUCUUGGCACUCACUGUGCC-ACUGUCGCU .-(-(.(((((..-...........(((((((((.....((.....))..(((((.....))))))))))))))((....))...))))).)).((((((.....)))))-)........ ( -29.60) >DroPer_CAF1 193883 118 - 1 --GAGUACACAGCUUGUCACCGCAUGACAGGCUCAGACAUGAAAUCUGUAUAAAUUUUGUGCAGACUGUACUUCCUUUACGGUUGUCAAACAGCUGGCACUCACUGUGUGUUCUGUCGCA --(((((((.(((((((((.....)))))))))...........((((((((.....)))))))).)))))))......((((((.....))))))((((.....))))........... ( -35.80) >consensus __GCAUACUCAAC_UGUCUUUCCGUGACAAACUAAAAUAUCAAAUGCGAAUAUAUUGUAAGCGAGCUGCGAUUCCUUCACGGUUGUCAGACUCUUGGCACUCACUGUGCC_ACCGUCUCU ........................((((((..............................((.....))....((.....))))))))(((....(((((.....)))))....)))... (-14.06 = -13.73 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:29 2006