
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,354,808 – 14,354,940 |
| Length | 132 |
| Max. P | 0.671206 |

| Location | 14,354,808 – 14,354,908 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 81.84 |
| Mean single sequence MFE | -22.35 |
| Consensus MFE | -16.84 |
| Energy contribution | -17.15 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
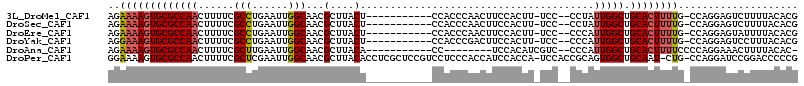
>3L_DroMel_CAF1 14354808 100 - 23771897 AGAAAAGUGCGCCAACUUUUCGCCUGAAUUGGCAACGCUUACU-----------CCACCCAACUUCCACUU-UCC--CCUAUUGGCUGCACUUUUG-CCAGGAGUCUUUUACACG .(((((((......)))))))(((......))).......(((-----------((...............-...--((....))..(((....))-)..))))).......... ( -21.00) >DroSec_CAF1 179325 100 - 1 AGAAAAGUGCGCCAACUUUUCGCCUGAAUUGGCAACGCUUACU-----------CCACCCAACUUCCACUU-UCC--CCUAUUGGCUGCACUUUUG-CCAGGAGUCUUUUACACG .(((((((......)))))))(((......))).......(((-----------((...............-...--((....))..(((....))-)..))))).......... ( -21.00) >DroEre_CAF1 177907 100 - 1 AGAAAAGUGCGCCAACUUUUCGCCUGAAUUGGCAACGCUUACU-----------CCACCCAACUUCCACUU-UCC--CCCAUUGGCUGCACUUUUG-CCAGGAGUAUUUUACACG .(((((((......)))))))(((......)))......((((-----------((..((((.........-...--....))))..(((....))-)..))))))......... ( -22.49) >DroYak_CAF1 185458 100 - 1 AGGAAAGUGCGCCAACUUUUCGCCUGAAUUGGCAACGCUUACU-----------CCACCCGACUUCCACUU-UCC--CCCAUUGGCUGCACUUUUG-CCAGGAGUCCUUUACACG .((((((((.(((((.(((......))))))))..)))))..)-----------))....((((((.....-...--.....((((.........)-)))))))))......... ( -22.31) >DroAna_CAF1 170953 93 - 1 AGAAAAGUGCGCCAACUUUUCGCUUGAAUUGGCAACGCUUACA-----------CC--------UCCACAUCGUC--CCCAUUGGCUGCACUUUUCCCCAGGAAACUUUUACAC- .((((((((((((((...(((....))).(((..(((......-----------..--------.......))).--.)))))))).)))))))))...((....)).......- ( -23.26) >DroPer_CAF1 189952 112 - 1 GGAAAAGUGCGCCAACUUUUCGCUCGAAUUGGCAACGCUUACACCUCGCUCCGUCCUCCCACCAUCCACCA-UCCACCGCAGUGGCUGCAAU-CUG-CCAGGAUCCGGACCCCCG ((..(((((.(((((...(((....))))))))..))))).........((((((((..((........((-.((((....)))).))....-.))-..))))..))))...)). ( -24.02) >consensus AGAAAAGUGCGCCAACUUUUCGCCUGAAUUGGCAACGCUUACU___________CCACCCAACUUCCACUU_UCC__CCCAUUGGCUGCACUUUUG_CCAGGAGUCUUUUACACG ..(((((((((((((......(((......)))...(....).......................................))))).)))))))).................... (-16.84 = -17.15 + 0.31)


| Location | 14,354,842 – 14,354,940 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.87 |
| Mean single sequence MFE | -22.07 |
| Consensus MFE | -16.00 |
| Energy contribution | -15.62 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
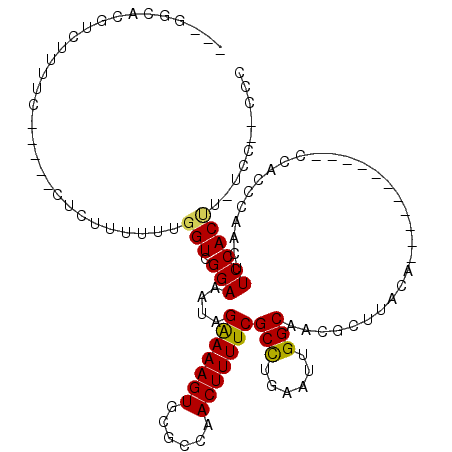
>3L_DroMel_CAF1 14354842 98 - 23771897 ---GGCACGUCUUUUC-----CUCUUUUUUGGUCGGAAAUAGAAAAGUGCGCCAACUUUUCGCCUGAAUUGGCAACGCUUACU-----------CCACCCAACUUCCACUU-UCC--CCU ---.............-----.......((((..(((....(((((((......)))))))(((......))).........)-----------))..)))).........-...--... ( -20.10) >DroPse_CAF1 175566 116 - 1 ---GGCACGUCUUCCCUGUCUUUAUUUUUUGGUCGGAAAUGGAAAAGUGCGCCAACUUUUCGCUCGAAUUGGCAACGCUUACACCUCGCUCCGUCCUCCCACCAUCCACCA-UCCACCGC ---((((.(.....).)))).........((((.(((.(((((...((..(((((...(((....)))))))).))((.........)))))))..))).)))).......-........ ( -23.00) >DroEre_CAF1 177941 97 - 1 ----GCACGUCUUUUC-----CUCCUUUUUGGUCGGAAAUAGAAAAGUGCGCCAACUUUUCGCCUGAAUUGGCAACGCUUACU-----------CCACCCAACUUCCACUU-UCC--CCC ----((((.(((((((-----(.((.....))..))))).)))...))))(((((.(((......))))))))..........-----------.................-...--... ( -21.90) >DroYak_CAF1 185492 101 - 1 GUGGGCACGUCUUUUC-----CUCUUUUUUGGUCGGAAAUAGGAAAGUGCGCCAACUUUUCGCCUGAAUUGGCAACGCUUACU-----------CCACCCGACUUCCACUU-UCC--CCC (((((...........-----.........((((((.....(((((((......)))))))(((......)))..........-----------....)))))))))))..-...--... ( -24.95) >DroAna_CAF1 170987 91 - 1 ---GGCACGUCUUUUC-----CUCUUUUUUGGUCGGAAAUAGAAAAGUGCGCCAACUUUUCGCUUGAAUUGGCAACGCUUACA-----------CC--------UCCACAUCGUC--CCC ---.((((.(((((((-----(.((.....))..))))).)))...))))(((((.(((......))))))))..........-----------..--------...........--... ( -19.50) >DroPer_CAF1 189985 116 - 1 ---GGCACGUCUUCCCUGUCUUUAUUUUUUGGUCGGAAAUGGAAAAGUGCGCCAACUUUUCGCUCGAAUUGGCAACGCUUACACCUCGCUCCGUCCUCCCACCAUCCACCA-UCCACCGC ---((((.(.....).)))).........((((.(((.(((((...((..(((((...(((....)))))))).))((.........)))))))..))).)))).......-........ ( -23.00) >consensus ___GGCACGUCUUUUC_____CUCUUUUUUGGUCGGAAAUAGAAAAGUGCGCCAACUUUUCGCCUGAAUUGGCAACGCUUACA___________CCACCCAACUUCCACUU_UCC__CCC ..............................(((.(((....(((((((......)))))))(((......)))...............................)))))).......... (-16.00 = -15.62 + -0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:27 2006