| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,316,865 – 14,316,996 |
| Length | 131 |
| Max. P | 0.717973 |

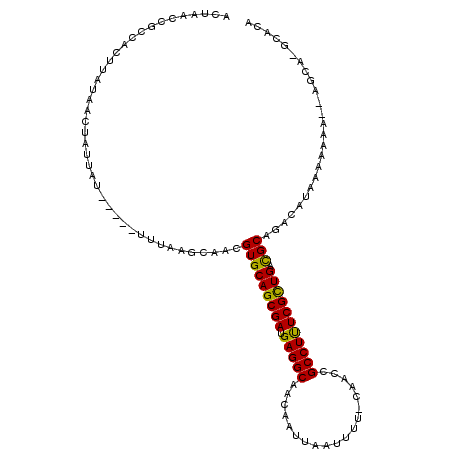
| Location | 14,316,865 – 14,316,964 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 86.30 |
| Mean single sequence MFE | -22.63 |
| Consensus MFE | -18.76 |
| Energy contribution | -18.26 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14316865 99 + 23771897 ACUAACCGCCACUUAUAACUAUUAU-----UUUAAGCAACGUGCAGCGAUGAGGCAACAAUUAAUUUU-CAACCGCCUUUCGCUGACGCAGACAUAAAAAAA--AGC------- .........................-----..........(((((((((.(((((.............-.....))))))))))).))).............--...------- ( -19.17) >DroSec_CAF1 141662 106 + 1 ACUAGCCGCCACUUAUAACUAUUAU-----UUUAAGCAACGUGCAGCGAUGAGGCAACAAUUAAUUUU-CAACCGCCUUUCGCUGACGCAGACAUGAAAAAA--AGCAGACACA .((.((.((.....(((.....)))-----.....))...(((((((((.(((((.............-.....))))))))))).))).............--.))))..... ( -20.97) >DroSim_CAF1 142313 106 + 1 ACUAGCCGCCACUUAUAACUAUUAU-----UUUAAGCAACGUGCAGCGAUGAGGCAACAAUUAAUUUG-CAACCGCCUUUCGCUGACGCAGACAUAAAAAAA--AGCAGGCACA ....(((((..((((..........-----..))))....(((((((((.(((((..(((.....)))-.....))))))))))).))).............--.)).)))... ( -26.20) >DroEre_CAF1 141953 113 + 1 ACUAACCGCCACUUAUAACUAUUAUGUUAUAUUAAGCAACGUGCAGCGAUGAGGCAACAAUUAUUUUU-CAACGGCCUUUCGCUGAUGCAGACAAAAAAAAACAAACAAGCACA .......((....((((((......))))))....))...(((((((((.(((((.............-.....))))))))))..((....))...............)))). ( -23.67) >DroYak_CAF1 146166 108 + 1 ACUAACCGCCACUUAUAACUAUUAU-----UUUAAGCAACGUGCAGCGAUGAGGCAACAAUUAUUUUU-CAACCGCCUUUCGCUGACGCAGACACAAAAAAACUAGCAAGCGCG ......(((..((((..........-----..))))....(((((((((.(((((.............-.....))))))))))).)))....................))).. ( -21.67) >DroAna_CAF1 135497 106 + 1 ACUAACCGCCACUUAUUACUAUUAU-----UUUAAGCAACGUGCAGCGAUGAGGCAACAAUUAUUGCUGGCACCGCCUCUCGUUGUUGCAGACACAACAAAAUU-GCA--CUCC .........................-----.....((((..((((((((((((((((......)))).(((...))).))))))))))))(......)....))-)).--.... ( -24.10) >consensus ACUAACCGCCACUUAUAACUAUUAU_____UUUAAGCAACGUGCAGCGAUGAGGCAACAAUUAAUUUU_CAACCGCCUUUCGCUGACGCAGACAUAAAAAAA__AGCA_GCACA ........................................(((((((((.(((((...................))))))))))).)))......................... (-18.76 = -18.26 + -0.50)


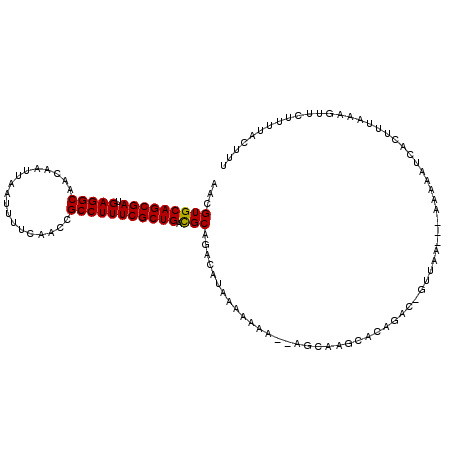
| Location | 14,316,897 – 14,316,996 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 84.06 |
| Mean single sequence MFE | -20.96 |
| Consensus MFE | -18.79 |
| Energy contribution | -18.63 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14316897 99 + 23771897 AACGUGCAGCGAUGAGGCAACAAUUAAUUUUCAACCGCCUUUCGCUGACGCAGACAUAAAAAAA--AGC-----------GUUAA----AAAAUCACUCUUAAGUUCUUUUACUUU ...(((((((((.(((((..................)))))))))))((((.............--.))-----------))...----.....)))....((((......)))). ( -19.71) >DroSec_CAF1 141694 110 + 1 AACGUGCAGCGAUGAGGCAACAAUUAAUUUUCAACCGCCUUUCGCUGACGCAGACAUGAAAAAA--AGCAGACACAGAC-GUUAA---AAAAAUCACUUUAAAGUUCUUUUACUUU ...(((((((((.(((((..................))))))))))).)))..........(((--((........(((-.((((---(........))))).))))))))..... ( -20.57) >DroSim_CAF1 142345 110 + 1 AACGUGCAGCGAUGAGGCAACAAUUAAUUUGCAACCGCCUUUCGCUGACGCAGACAUAAAAAAA--AGCAGGCACAGAC-GUUAA---AAAAAUCACUUUAAAGUUCUUUUACUUU ...(((((((((.(((((..(((.....))).....))))))))))).)))..........(((--((........(((-.((((---(........))))).))))))))..... ( -21.00) >DroEre_CAF1 141990 116 + 1 AACGUGCAGCGAUGAGGCAACAAUUAUUUUUCAACGGCCUUUCGCUGAUGCAGACAAAAAAAAACAAACAAGCACAGACAGUUAAAAAAAAAAUUACUUAUAUACUUUUUCACUUU ...(((((((((.(((((..................))))))))))..((....))...............))))......................................... ( -17.17) >DroYak_CAF1 146198 116 + 1 AACGUGCAGCGAUGAGGCAACAAUUAUUUUUCAACCGCCUUUCGCUGACGCAGACACAAAAAAACUAGCAAGCGCGGACAGUUAAAGAAAAAAUCAUUUAAAAGUUUUUUUAUUUC ...(((((((((.(((((..................))))))))))).)))......(((((((((.((....))......(((((((.....)).))))).)))))))))..... ( -26.37) >consensus AACGUGCAGCGAUGAGGCAACAAUUAAUUUUCAACCGCCUUUCGCUGACGCAGACAUAAAAAAA__AGCAAGCACAGAC_GUUAA___AAAAAUCACUUUAAAGUUCUUUUACUUU ...(((((((((.(((((..................))))))))))).)))................................................................. (-18.79 = -18.63 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:16 2006