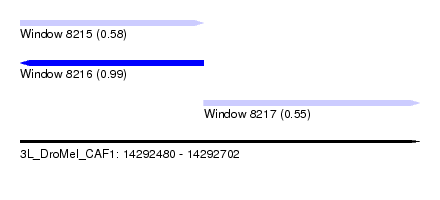
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,292,480 – 14,292,702 |
| Length | 222 |
| Max. P | 0.985470 |

| Location | 14,292,480 – 14,292,582 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.42 |
| Mean single sequence MFE | -21.50 |
| Consensus MFE | -14.90 |
| Energy contribution | -14.57 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14292480 102 + 23771897 GGGUCUGACUAUGCAAAAAUGCAGCAGCCAAAAAGCACAUAAAAUUAUUAUGUGACGAGGAACGUGGGAUGCGAAUAUAUGGUGGUAGU-GGGG-----------------GGAAAAGCG ...(((.((((((((....))).(((.((......(((((((.....)))))))(((.....))).)).)))............)))))-.)))-----------------......... ( -22.50) >DroSec_CAF1 117728 101 + 1 GGGUCUGACUAUGCAAAAAUGCAGCAGCCAAAAAGCACAUAAAAUUAUUAUGUGACGAGGAACGUGGGAUGCGAAAAUC-GGGGGUUGU-GGGG-----------------GGAAAAGGG ...(((..((.((((....))))((((((......(((((((.....))))))).(((....(((.....)))....))-)..))))))-))..-----------------)))...... ( -25.30) >DroSim_CAF1 117829 101 + 1 GGGUCUGACUAUGCAAAAAUGCAGCAGCCAAAAAGCACAUAAAAUUAUUAUGUGACGAGGAACGUGGGAUGCGAAAAUC-GGGGGUUGU-GGGG-----------------CGAAAAGGG ..((((.....((((....))))((((((......(((((((.....))))))).(((....(((.....)))....))-)..))))))-.)))-----------------)........ ( -25.70) >DroEre_CAF1 117861 100 + 1 GGGUCUGACUAUGCAAAAAUGCAGCAGCCAAAAAGCACAUAAAAUUAUUAUGUGACGAGGAACGUGGGGAGUGAAAAUC-GGGGGUCGU-GGGGAAGGG------------------GGG ...(((..((.((((....))))((.(((......(((((((.....))))))).(((...((.......)).....))-)..))).))-.))..))).------------------... ( -19.10) >DroYak_CAF1 121617 119 + 1 GGGUCUGACUAUGCAAAAAUGCAGCAGCCAAAAAGCACAUAAAAUUAUUAUGUGACGAGGAACGUGGGGAGCGAAAAUC-GGUGGUCGUUGGGGAAGGGAAAGGGAAAGGGGAAAAAGCG ...(((..((...(.....(.((((.((((.....(((((((.....))))))).(((....(((.....)))....))-).)))).)))).).....)....))..))).......... ( -22.30) >DroAna_CAF1 112730 85 + 1 AAGUCUGACUAUGCAAAAAUGCAGCAGCCAAAAAGCAUAUAAAAUUAUUAUGUGACGAGGAACGUGGGGAAUGAGCACC-AGGA----------------------------------GG ..(.(((....((((....)))).))).).....((((((((.....))))))).)......(.(((..........))-).).----------------------------------.. ( -14.10) >consensus GGGUCUGACUAUGCAAAAAUGCAGCAGCCAAAAAGCACAUAAAAUUAUUAUGUGACGAGGAACGUGGGAAGCGAAAAUC_GGGGGUCGU_GGGG_________________GGAAAAGGG .((.(((....((((....)))).))))).....((((((((.....))))))).)......(((.....)))............................................... (-14.90 = -14.57 + -0.33)


| Location | 14,292,480 – 14,292,582 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.42 |
| Mean single sequence MFE | -17.78 |
| Consensus MFE | -17.42 |
| Energy contribution | -17.58 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14292480 102 - 23771897 CGCUUUUCC-----------------CCCC-ACUACCACCAUAUAUUCGCAUCCCACGUUCCUCGUCACAUAAUAAUUUUAUGUGCUUUUUGGCUGCUGCAUUUUUGCAUAGUCAGACCC .........-----------------....-.................................(.(((((((.....))))))))..((((((((.((((....))))))))))))... ( -18.30) >DroSec_CAF1 117728 101 - 1 CCCUUUUCC-----------------CCCC-ACAACCCCC-GAUUUUCGCAUCCCACGUUCCUCGUCACAUAAUAAUUUUAUGUGCUUUUUGGCUGCUGCAUUUUUGCAUAGUCAGACCC .........-----------------....-........(-((....((.......))....))).(((((((.....)))))))...((((((((.((((....))))))))))))... ( -18.70) >DroSim_CAF1 117829 101 - 1 CCCUUUUCG-----------------CCCC-ACAACCCCC-GAUUUUCGCAUCCCACGUUCCUCGUCACAUAAUAAUUUUAUGUGCUUUUUGGCUGCUGCAUUUUUGCAUAGUCAGACCC .........-----------------....-........(-((....((.......))....))).(((((((.....)))))))...((((((((.((((....))))))))))))... ( -18.70) >DroEre_CAF1 117861 100 - 1 CCC------------------CCCUUCCCC-ACGACCCCC-GAUUUUCACUCCCCACGUUCCUCGUCACAUAAUAAUUUUAUGUGCUUUUUGGCUGCUGCAUUUUUGCAUAGUCAGACCC ...------------------.........-.........-.......................(.(((((((.....))))))))..((((((((.((((....))))))))))))... ( -18.30) >DroYak_CAF1 121617 119 - 1 CGCUUUUUCCCCUUUCCCUUUCCCUUCCCCAACGACCACC-GAUUUUCGCUCCCCACGUUCCUCGUCACAUAAUAAUUUUAUGUGCUUUUUGGCUGCUGCAUUUUUGCAUAGUCAGACCC ..............................((((.....(-(.....)).......))))....(.(((((((.....))))))))..((((((((.((((....))))))))))))... ( -18.90) >DroAna_CAF1 112730 85 - 1 CC----------------------------------UCCU-GGUGCUCAUUCCCCACGUUCCUCGUCACAUAAUAAUUUUAUAUGCUUUUUGGCUGCUGCAUUUUUGCAUAGUCAGACUU ..----------------------------------....-((.((...........)).))..(((..((((.....))))........((((((.((((....))))))))))))).. ( -13.80) >consensus CCCUUUUCC_________________CCCC_ACAACCCCC_GAUUUUCGCACCCCACGUUCCUCGUCACAUAAUAAUUUUAUGUGCUUUUUGGCUGCUGCAUUUUUGCAUAGUCAGACCC ................................................................(.(((((((.....))))))))..((((((((.((((....))))))))))))... (-17.42 = -17.58 + 0.17)



| Location | 14,292,582 – 14,292,702 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.28 |
| Mean single sequence MFE | -30.33 |
| Consensus MFE | -28.71 |
| Energy contribution | -28.68 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.09 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14292582 120 + 23771897 GAGGACAAUGCUUAAAGGCAUUUCGGACUCGGCCAAAAAUCAGACUCAGCAUGUGCGCAUAAAGCUCAAAUAAACUCAAGUUGUUGACAAAGGGGUUAAAGGCCAUAAUUGUGGCCGUCG (((..(((((((....))))))..)..)))(((.........(((((.((....))((.....))((((...(((....))).)))).....)))))...((((((....))))))))). ( -32.30) >DroSec_CAF1 117829 120 + 1 GAGGACAAUGCUCAAAGGCAUUUCGGACUCGGCCAAAAAUCAGACUCAGCAUGUGCGCAUAAAGCUCAAAUAAACUCAAGUUGUUGACAAAGGGGUUAAAGGCCAUAAUUGUGGCCGUCG (((..(((((((....))))))..)..)))(((.........(((((.((....))((.....))((((...(((....))).)))).....)))))...((((((....))))))))). ( -31.90) >DroSim_CAF1 117930 120 + 1 GAGGACAAUGCUCAAAGGCAUUUCGGACUCGGCCAAAAAUCAGACUCAGCAUGUGCGCAUAAAGCUCAAAUAAACUCAAGUUGUUGACAAAGGGGUUAAAGGCCAUAAUUGUGGCCGUCG (((..(((((((....))))))..)..)))(((.........(((((.((....))((.....))((((...(((....))).)))).....)))))...((((((....))))))))). ( -31.90) >DroEre_CAF1 117961 120 + 1 UAGGACAAUGCUCAAAGGCAUUUCGGACUCGGCCAAAAAUCAGACUCAGCAUGUGCGCAUAAAGCUCAAAUAAACUCAAGUUGUUGACAAAGGGGUUAAAGGCCAUAAUUGUGGCCGUCG ...((.((((((....))))))))((......))........(((......((.((.......)).))....(((((...(((....)))..)))))...((((((....))))))))). ( -28.30) >DroYak_CAF1 121736 120 + 1 AGGGACAACGCUCAAAGGCAUUUUGGACUUGGCCAAAAAUCAGACUCAGCAUGUGCGCAUAAAGCUCAAAUAAACUCAAGUUGUUGACAAAGGGGUUAAAGGCCAUAAUUGUGGCCGUCG ...(((...(((.....((.((((((......))))))..........((....))))....))).......(((((...(((....)))..)))))...((((((....))))))))). ( -30.70) >DroAna_CAF1 112815 120 + 1 AGGGACAAUGCUCAAAGGCAUUUCGGACUCGGCCAAAAAUCAGACUCAGCAUGUGCACAUAAAGCUCAAAUAAACUCAAGUUGUUGACAAAGGGGUUAAAGGCCAUAAUUGUGGCCGUUG ...((.((((((....))))))))((......))............((((.((.((.......)).))....(((((...(((....)))..)))))...((((((....)))))))))) ( -26.90) >consensus GAGGACAAUGCUCAAAGGCAUUUCGGACUCGGCCAAAAAUCAGACUCAGCAUGUGCGCAUAAAGCUCAAAUAAACUCAAGUUGUUGACAAAGGGGUUAAAGGCCAUAAUUGUGGCCGUCG (((..(((((((....))))))..)..)))(((.........(((((.((....)).......(.((((...(((....))).)))))....)))))...((((((....))))))))). (-28.71 = -28.68 + -0.03)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:09 2006