
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,272,366 – 14,272,496 |
| Length | 130 |
| Max. P | 0.700289 |

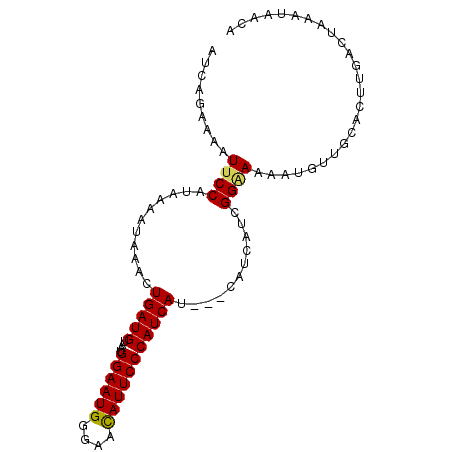
| Location | 14,272,366 – 14,272,457 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 93.98 |
| Mean single sequence MFE | -15.38 |
| Consensus MFE | -12.47 |
| Energy contribution | -12.27 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14272366 91 - 23771897 AUCAGAAAAUUCCAUAAAAUAAACUGAUGUAAUGGAAUGUUCCCAUUCCCAUCAU---CAUCAUCGGGAAAAUGUUGCACUUGACUAAAUAACA .((((((.(((((((...((......))...))))))).)))...(((((.....---.......)))))...........))).......... ( -13.50) >DroSec_CAF1 102321 94 - 1 AUCAGAAAAUUCCAUAAAAUAAACUGAUGUAAUGGAAUGGGAACAUUCCCAUCAUCAUCAUCAUCGGAAAAAUGUUGCACUUGACUAAAUAACA .......................((((((..((((.((((((....))))))..))))...))))))........................... ( -16.40) >DroSim_CAF1 102412 94 - 1 AUCAGAAAAUUCCAUAAAAUAAACUGAUGUAAUGGAAUGGGAACAUUCCCAUCAUCAUCAUCAUCGGAAAAAUGUUGCACUUGACUAAAUAACA .......................((((((..((((.((((((....))))))..))))...))))))........................... ( -16.40) >DroEre_CAF1 102100 91 - 1 AUCAGAGAAUUCCAUAAAAUAAACUGAUGUAAUGGAAUGGGAAUAUUCCCAUCAU---CAUCAUCGGGAAAAUGUUGCACUUGACUAAAUAACA .......................((((((.....((.(((((....)))))))..---...))))))........................... ( -15.70) >DroYak_CAF1 105870 91 - 1 AUCAGAGAAUUCCAUAAAAUAAACUGAUGUAAUGGAAUGGGCAUAUUCCCAUCAU---CAUCAUCGGCAAAAUGUUGCACUUGACUAAAUAACA ........................(((((....((((((....))))))...)))---))(((...((((....))))...))).......... ( -14.90) >consensus AUCAGAAAAUUCCAUAAAAUAAACUGAUGUAAUGGAAUGGGAACAUUCCCAUCAU___CAUCAUCGGAAAAAUGUUGCACUUGACUAAAUAACA .........((((...........(((((....((((((....)))))))))))...........))))......................... (-12.47 = -12.27 + -0.20)

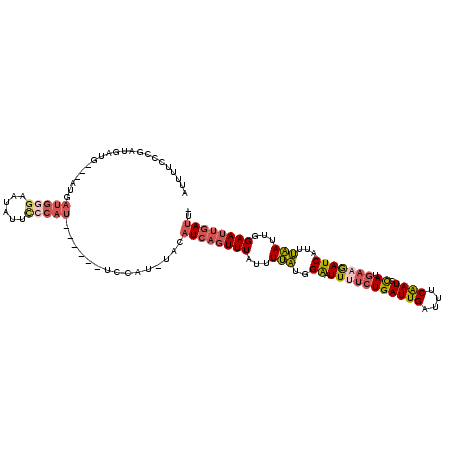
| Location | 14,272,387 – 14,272,496 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.02 |
| Mean single sequence MFE | -25.10 |
| Consensus MFE | -11.73 |
| Energy contribution | -12.85 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.680520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
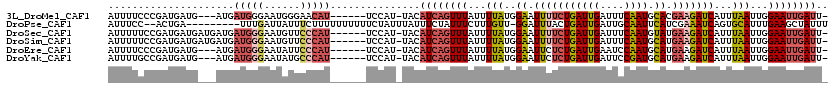
>3L_DroMel_CAF1 14272387 109 + 23771897 AUUUUCCCGAUGAUG---AUGAUGGGAAUGGGAACAU------UCCAU-UACAUCAGUUUAUUUUAUGGAAUUUUCUGAUUGAUUUCAAUGCACGAAGAUCAUUUAAUUGGAAUUGAUU- ...((((.(((((((---(((...((((((....)))------)))..-..)))))..)))))....))))......(((..((((((((................))))))))..)))- ( -26.29) >DroPse_CAF1 99352 108 + 1 AUUUCC--ACUGA---------UUUGAUUAUUUCUUUUUUUUUUCUAUUUAUUUCUAUUUCUUUGUU-GGAUUUACUGAUUGAUUGCAAUUCAUCGAAAUCAGUGCAUUUGAAGCUAUUU .....(--(((((---------((((((.....................((..((((.........)-)))..))..(((((....))))).))).)))))))))............... ( -13.60) >DroSec_CAF1 102342 112 + 1 AUUUUUCCGAUGAUGAUGAUGAUGGGAAUGUUCCCAU------UCCAU-UACAUCAGUUUAUUUUAUGGAAUUUUCUGAUUGAUUUCAAUGUAUGAAGAUCAUUUAAUUGGAAUUGAUU- ....(((((.(((((.(((((((((((....))))))------..)))-)))))))..........)))))......(((..((((((((..(((.....)))...))))))))..)))- ( -29.30) >DroSim_CAF1 102433 112 + 1 AUUUUUCCGAUGAUGAUGAUGAUGGGAAUGUUCCCAU------UCCAU-UACAUCAGUUUAUUUUAUGGAAUUUUCUGAUUGAUUUCAAUGCAUGAAGAUCAUUUAAUUGGAAUUGAUU- ....(((((.(((((.(((((((((((....))))))------..)))-)))))))..........)))))......(((..((((((((..(((.....)))...))))))))..)))- ( -28.60) >DroEre_CAF1 102121 109 + 1 AUUUUCCCGAUGAUG---AUGAUGGGAAUAUUCCCAU------UCCAU-UACAUCAGUUUAUUUUAUGGAAUUCUCUGAUUGAAUCCAAUGCAUGAAGAUCAUUUAAUUGGAAUUGAUU- ........((((.((---(((((((((....))))))------..)))-))))))......................(((..(.((((((..(((.....)))...)))))).)..)))- ( -25.30) >DroYak_CAF1 105891 109 + 1 AUUUUGCCGAUGAUG---AUGAUGGGAAUAUGCCCAU------UCCAU-UACAUCAGUUUAUUUUAUGGAAUUCUCUGAUUGAUUCCGAUGCAUGAAGAUCAUUUAAUUGGAAUUGAUU- ........((((.((---((((((((......)))))------..)))-))))))......................(((..((((((((..(((.....)))...))))))))..)))- ( -27.50) >consensus AUUUUCCCGAUGAUG___AUGAUGGGAAUAUUCCCAU______UCCAU_UACAUCAGUUUAUUUUAUGGAAUUUUCUGAUUGAUUUCAAUGCAUGAAGAUCAUUUAAUUGGAAUUGAUU_ .....................(((((......)))))...............((((((((...(((..((.(((((((((((....)))).)).)))))))...)))...)))))))).. (-11.73 = -12.85 + 1.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:04 2006