| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,271,927 – 14,272,021 |
| Length | 94 |
| Max. P | 0.997880 |

| Location | 14,271,927 – 14,272,021 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 73.24 |
| Mean single sequence MFE | -33.72 |
| Consensus MFE | -15.86 |
| Energy contribution | -16.05 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.47 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14271927 94 + 23771897 AGGA--------CCUUC-AUUUGGGUGGAUGGC--GGUCCUGUUGUCUGGGGGGUGGAAUUCACCCCUUGAGAACAUUGUCACUCAUAUGGGGACAUGUUUUGCA ((((--------((.((-((((....)))))).--))))))(((.((.((((((((.....)))))))))).)))..((((.(((....)))))))......... ( -37.90) >DroSec_CAF1 101891 93 + 1 AGGA--------CCUUU-UUUGGGGGGGAUGGG--UGUCCUGUUGUCUGGG-GGUGCAAUUCACCCCUUGAGAACAUUGUCACUCAUAUGGGGAUAUGUUUUGCA ....--------((((.-..((((.(((((...--.)))))(((.((.(((-((((.....))))))).)).))).......))))...))))............ ( -31.00) >DroSim_CAF1 101973 94 + 1 AGGA--------CCUUUUUUUGGGGGGGAUUGG--GGUCCUGUUGUCUGGG-GGUGGAAUUCACCCCUUGAGAACAUUGUCACUCAUAUGGGGACAUGUUUUGCA ((((--------((((..(((.....)))..))--))))))(((.((.(((-((((.....))))))).)).)))..((((.(((....)))))))......... ( -36.50) >DroEre_CAF1 101665 92 + 1 AGCA--------UCUUUUUUUG-GG-GGAUGGG--GGCUCUGUUAUCUGGG-GGUGAAAUUCACCCCUUGAGAACAUUGUCACUCAUAUGGGGACAUGUUUUGCA .(((--------(((((.....-))-))))(((--(((..((((.((.(((-((((.....))))))).)).))))..))).)))(((((....)))))...)). ( -34.90) >DroYak_CAF1 105442 78 + 1 AGGA--------CCUCUUUUUUGGG-AGG-----------------UUGGG-GGUGAGAUUCACCCCCUGAGAACAUUGUCACUCAUAUGGGGACAUGUUUUGCA ..((--------((((((....)))-)))-----------------))(((-((((.....)))))))..(((((((.(((.(((....)))))))))))))... ( -35.90) >DroAna_CAF1 96908 102 + 1 GGGGUAGUUCUAUUUUUUUUUUUGUUGGAUGGGAGGAUCCUGUU--CGGAA-GGUGGCAGGCAGCCUCGGGGAAAAAUUUCACUCACAGGACUUGAUGUUUUGCA (((((.(((((((((((........((((((((.....))))))--)))))-)))))..))).)))))((((((....))).))).((((((.....)))))).. ( -26.10) >consensus AGGA________CCUUUUUUUGGGG_GGAUGGG__GGUCCUGUUGUCUGGG_GGUGGAAUUCACCCCUUGAGAACAUUGUCACUCAUAUGGGGACAUGUUUUGCA ..........................((((......))))......(.(((.((((.....))))))).)((((((.((((.(((....)))))))))))))... (-15.86 = -16.05 + 0.19)

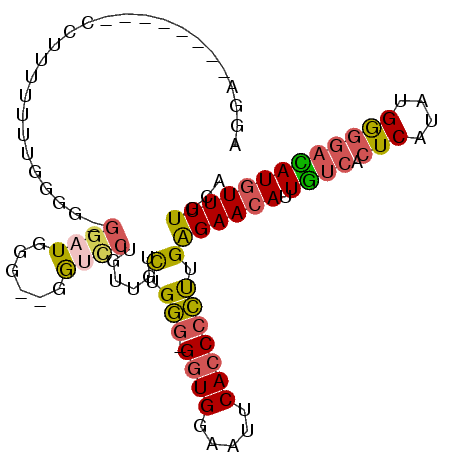

| Location | 14,271,927 – 14,272,021 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 73.24 |
| Mean single sequence MFE | -22.50 |
| Consensus MFE | -7.15 |
| Energy contribution | -8.32 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.32 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972155 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14271927 94 - 23771897 UGCAAAACAUGUCCCCAUAUGAGUGACAAUGUUCUCAAGGGGUGAAUUCCACCCCCCAGACAACAGGACC--GCCAUCCACCCAAAU-GAAGG--------UCCU .........((((......((((.(((...))))))).((((((.....))))))...))))..((((((--..(((........))-)..))--------)))) ( -27.70) >DroSec_CAF1 101891 93 - 1 UGCAAAACAUAUCCCCAUAUGAGUGACAAUGUUCUCAAGGGGUGAAUUGCACC-CCCAGACAACAGGACA--CCCAUCCCCCCCAAA-AAAGG--------UCCU .......(((((....)))))((.(((..((((.((..((((((.....))))-))..)).))))(((..--....)))........-....)--------)))) ( -21.50) >DroSim_CAF1 101973 94 - 1 UGCAAAACAUGUCCCCAUAUGAGUGACAAUGUUCUCAAGGGGUGAAUUCCACC-CCCAGACAACAGGACC--CCAAUCCCCCCCAAAAAAAGG--------UCCU .........(((((.(....).).)))).((((.((..((((((.....))))-))..)).))))(((((--...................))--------))). ( -24.51) >DroEre_CAF1 101665 92 - 1 UGCAAAACAUGUCCCCAUAUGAGUGACAAUGUUCUCAAGGGGUGAAUUUCACC-CCCAGAUAACAGAGCC--CCCAUCC-CC-CAAAAAAAGA--------UGCU .(((.....(((((.(....).).)))).((((.((..((((((.....))))-))..)).)))).....--.......-..-..........--------))). ( -21.40) >DroYak_CAF1 105442 78 - 1 UGCAAAACAUGUCCCCAUAUGAGUGACAAUGUUCUCAGGGGGUGAAUCUCACC-CCCAA-----------------CCU-CCCAAAAAAGAGG--------UCCU .....(((((((((.(....).).))).)))))....(((((((.....))))-))).(-----------------(((-(........))))--------)... ( -25.70) >DroAna_CAF1 96908 102 - 1 UGCAAAACAUCAAGUCCUGUGAGUGAAAUUUUUCCCCGAGGCUGCCUGCCACC-UUCCG--AACAGGAUCCUCCCAUCCAACAAAAAAAAAAAUAGAACUACCCC .............(((((((..(.(((.....((...))(((.....)))...-)))).--.))))))).................................... ( -14.20) >consensus UGCAAAACAUGUCCCCAUAUGAGUGACAAUGUUCUCAAGGGGUGAAUUCCACC_CCCAGACAACAGGACC__CCCAUCC_CCCAAAAAAAAGG________UCCU .......(((((....)))))........((((.((..((((((.....)))).))..)).))))........................................ ( -7.15 = -8.32 + 1.17)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:03 2006