
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,267,266 – 14,267,439 |
| Length | 173 |
| Max. P | 0.927402 |

| Location | 14,267,266 – 14,267,363 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 92.36 |
| Mean single sequence MFE | -30.68 |
| Consensus MFE | -24.36 |
| Energy contribution | -24.56 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14267266 97 + 23771897 UAG---------CCACUAUAGUGACUCUUGCAGGGCAGUUUAGUCGCUGUCCGGUUCGUGUUUUACUGUUUCCACAGCUUUUAGCCGCUGCUGACGGUCGGAUGCU .((---------((...(((((((((.((((...))))...)))))))))..)))).(((((..((((((....((((........))))..))))))..))))). ( -29.90) >DroSec_CAF1 97271 97 + 1 UAG---------UCACUAUAGUGACUCUUGCAGGGCAGUUUAGUCGCUGUCCGGUUCUUGUUUUACUGUUUCCACAGCUUUUAGCCGCUGCUGACGGUCGGAUGCU .((---------((((....))))))...((.(((((((......))))))).......(((..((((((....((((........))))..))))))..))))). ( -31.20) >DroSim_CAF1 97340 97 + 1 UAG---------UCACUAUAGUGACUCUUGCAGGGCAGUUUAGUCGCUGUCCGGUUCUUGUUUUACUGUUUCCACAGCUUUUAGCCGCUGCUGACGUUCGGAUGCU .((---------((((....))))))...((((((((((......))))))).............(((....((((((........)))).)).....))).))). ( -27.00) >DroEre_CAF1 96827 102 + 1 UAGUCACUA----CACUAUAGUGACUCUUGCAGGGCAGUUUAGUCGCUGUCCGGUUCUUGUUUUACUGUUUCCACAGCUUUUAGCUGCUGCUGAUGGUCGGAUGCU .((((((((----.....))))))))...((.(((((((......))))))).......(((..((((((..((((((.....)))).))..))))))..))))). ( -33.30) >DroYak_CAF1 100525 105 + 1 UAA-GACUAUAGUCACUAUAGUGACUCUUGCAGGGCAGUUUAGUCGCUGUCCGGUUCUUGUUUUACUGUUUCCACAGCUUUUAGCUGCUGCUGAUGGUCGGAUGCU .((-((((..((((((....))))))......(((((((......))))))))).))))(((..((((((..((((((.....)))).))..))))))..)))... ( -32.00) >consensus UAG_________UCACUAUAGUGACUCUUGCAGGGCAGUUUAGUCGCUGUCCGGUUCUUGUUUUACUGUUUCCACAGCUUUUAGCCGCUGCUGACGGUCGGAUGCU .................(((((((..(..((.(((((((......))))))).))....)..))))))).(((...(((.(((((....))))).))).))).... (-24.36 = -24.56 + 0.20)


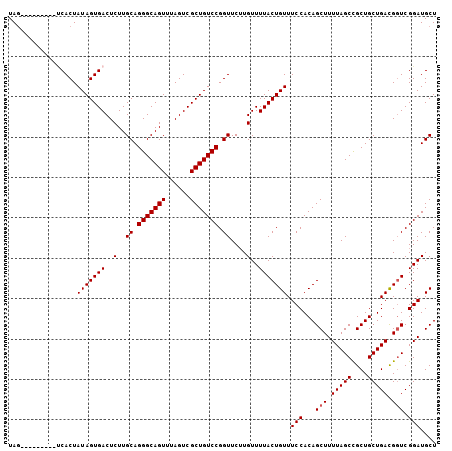
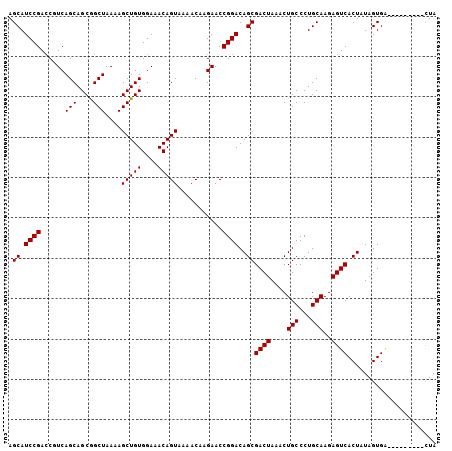
| Location | 14,267,266 – 14,267,363 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 92.36 |
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -21.04 |
| Energy contribution | -21.04 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14267266 97 - 23771897 AGCAUCCGACCGUCAGCAGCGGCUAAAAGCUGUGGAAACAGUAAAACACGAACCGGACAGCGACUAAACUGCCCUGCAAGAGUCACUAUAGUGG---------CUA .(((((((.((((.....))))......(((((....)))))...........))))(((........)))...)))...((((((....))))---------)). ( -26.30) >DroSec_CAF1 97271 97 - 1 AGCAUCCGACCGUCAGCAGCGGCUAAAAGCUGUGGAAACAGUAAAACAAGAACCGGACAGCGACUAAACUGCCCUGCAAGAGUCACUAUAGUGA---------CUA .(((((((.((((.....))))......(((((....)))))...........))))(((........)))...)))...((((((....))))---------)). ( -26.90) >DroSim_CAF1 97340 97 - 1 AGCAUCCGAACGUCAGCAGCGGCUAAAAGCUGUGGAAACAGUAAAACAAGAACCGGACAGCGACUAAACUGCCCUGCAAGAGUCACUAUAGUGA---------CUA ...............((((.(((.....(((((....)))))...........((.....))........)))))))...((((((....))))---------)). ( -26.30) >DroEre_CAF1 96827 102 - 1 AGCAUCCGACCAUCAGCAGCAGCUAAAAGCUGUGGAAACAGUAAAACAAGAACCGGACAGCGACUAAACUGCCCUGCAAGAGUCACUAUAGUG----UAGUGACUA .(((((((......(((....)))....(((((....)))))...........))))(((........)))...)))...(((((((((...)----)))))))). ( -27.60) >DroYak_CAF1 100525 105 - 1 AGCAUCCGACCAUCAGCAGCAGCUAAAAGCUGUGGAAACAGUAAAACAAGAACCGGACAGCGACUAAACUGCCCUGCAAGAGUCACUAUAGUGACUAUAGUC-UUA .((.((((......(((....)))....(((((....)))))...........))))..))(((((...(((...)))..((((((....)))))).)))))-... ( -29.30) >consensus AGCAUCCGACCGUCAGCAGCGGCUAAAAGCUGUGGAAACAGUAAAACAAGAACCGGACAGCGACUAAACUGCCCUGCAAGAGUCACUAUAGUGA_________CUA .((.((((......(((....)))....(((((....)))))...........))))..))((((....(((...)))..))))...................... (-21.04 = -21.04 + -0.00)

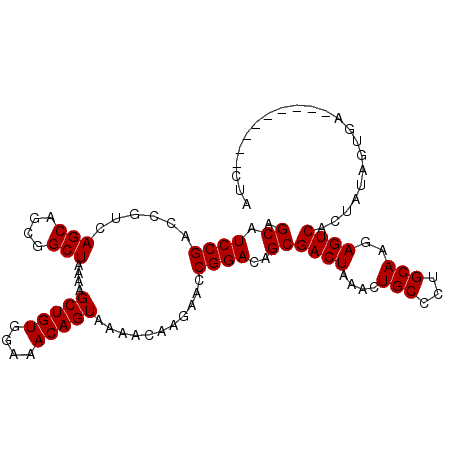
| Location | 14,267,294 – 14,267,399 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 90.63 |
| Mean single sequence MFE | -35.62 |
| Consensus MFE | -31.42 |
| Energy contribution | -32.03 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14267294 105 + 23771897 GUUUAGUCGC-UGUCCGGUUCGUGUUUUACUGUUUCCACAGCUUUUAGCCGCUGCUGACGGUCGGAUGCUCACGCGGCCGGCAUGAAAACUUAUGUCGGCGGCCUU .....(((((-((.(((((..(((((..((((((....((((........))))..))))))..)))))..)).))).)(((((((....)))))))))))))... ( -35.60) >DroSec_CAF1 97299 105 + 1 GUUUAGUCGC-UGUCCGGUUCUUGUUUUACUGUUUCCACAGCUUUUAGCCGCUGCUGACGGUCGGAUGCUCACGCGGCCGGCAUGAAAACUUAUGUCGGCGGCCUU ........((-.(((((...........((((((....((((........))))..)))))))))))))....((.((((((((((....)))))))))).))... ( -35.50) >DroSim_CAF1 97368 105 + 1 GUUUAGUCGC-UGUCCGGUUCUUGUUUUACUGUUUCCACAGCUUUUAGCCGCUGCUGACGUUCGGAUGCUCACGCGGCCGGCAUGAAAACUUAUGUCGGCGGCCUU ........((-.((((((...........((((....))))...(((((....)))))...))))))))....((.((((((((((....)))))))))).))... ( -33.60) >DroEre_CAF1 96860 105 + 1 GUUUAGUCGC-UGUCCGGUUCUUGUUUUACUGUUUCCACAGCUUUUAGCUGCUGCUGAUGGUCGGAUGCUCACGCGGCCGGCAUGAAAACUUAUGUCGGCGGCCUU ........((-.((((((...........((((....))))((.(((((....))))).))))))))))....((.((((((((((....)))))))))).))... ( -35.80) >DroYak_CAF1 100561 105 + 1 GUUUAGUCGC-UGUCCGGUUCUUGUUUUACUGUUUCCACAGCUUUUAGCUGCUGCUGAUGGUCGGAUGCUCACGCGGCCGGCAUGAAAACUUAUGUCGGCGGCCUU ........((-.((((((...........((((....))))((.(((((....))))).))))))))))....((.((((((((((....)))))))))).))... ( -35.80) >DroAna_CAF1 92692 89 + 1 GUUUAGACGCCUGUCCGACUUU-----------------GGCUUUUCGCUGCUGCUGAUGGUCGGAUGCUCACGCGGCCGGCAUGAAAACUUAUGUCGGCGGCCUU .....((.((..(((((((((.-----------------.((...........))..).))))))))))))..((.((((((((((....)))))))))).))... ( -37.40) >consensus GUUUAGUCGC_UGUCCGGUUCUUGUUUUACUGUUUCCACAGCUUUUAGCCGCUGCUGACGGUCGGAUGCUCACGCGGCCGGCAUGAAAACUUAUGUCGGCGGCCUU ........((..((((((...........((((....))))((.(((((....))))).))))))))))....((.((((((((((....)))))))))).))... (-31.42 = -32.03 + 0.61)

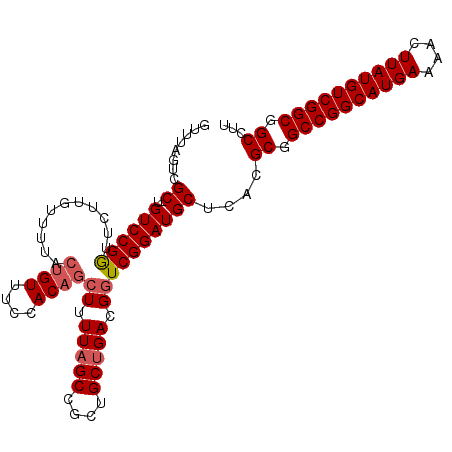

| Location | 14,267,329 – 14,267,439 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 84.00 |
| Mean single sequence MFE | -32.07 |
| Consensus MFE | -26.55 |
| Energy contribution | -26.05 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14267329 110 + 23771897 CACAGCUUUUAGCCGCUG---CUGAC---GGUCGGAUGCUCACGCGGCCGGCAUGAAAACUUAUGUCGGCGGCCUUUGGGCGCAAUUAUUUAUACAUGCCUCAUAAAAAUCGCAAG ...(((.((..(((((..---..).)---)))..)).)))...(((((((((((((....)))))))))).(((....))).............................)))... ( -34.70) >DroVir_CAF1 109384 100 + 1 -----GUUUUAGUUGUUG---CUGUU---GUUC----GUCCACGCGGCCGGCAUGAAAACUUAUGUCGGCGGCCUUUGGGCGCAAUUAUUUAUACAUGCCUCAUAAAAAUCG-AAA -----.(((((..((..(---((((.---((((----(((((.((.((((((((((....)))))))))).))...)))))).))).......))).))..)))))))....-... ( -30.90) >DroPse_CAF1 93833 97 + 1 -------------UGCUG---CUCCU---GGUCGGAUGCUCACGCGGCCGGCAUGAAAACUUAUGUCGGCGGCCUUUGGGCGCAAUUAUUUAUACAUGCCUCAUAAAAAUCGCAAG -------------(((((---((((.---....))).(((((.((.((((((((((....)))))))))).))...))))))))....(((((.........)))))....))).. ( -31.60) >DroGri_CAF1 119798 106 + 1 -----AUUUUAGUUGUUGUUGCUGCCGUUGUUC----GUCCACGCGGCCGGCAUGAAAACUUAUGUCGGCGGCCUUUGGGCGCAAUUAUUUAUACAUGCCUCAUAAAAAUCG-AAA -----.((((((..(((((.......((((..(----(((((.((.((((((((((....)))))))))).))...)))))))))).......))).))..).)))))....-... ( -31.04) >DroMoj_CAF1 131422 103 + 1 -----GUUUUAGUUGUUG---CUGUUGGUGUUC----GUCCACGCGGCCGGCAUGAAAACUUAUGUCGGCGGCCUUUGGGCGCAAUUAUUUAUACAUGCCUCAUAAAAAUCG-AAA -----.(((((..((..(---((((.(((((((----(((((.((.((((((((((....)))))))))).))...)))))).)).))))...))).))..)))))))....-... ( -32.60) >DroPer_CAF1 105854 97 + 1 -------------UGCUG---CUCCU---GGUCGGAUGCUCACGCGGCCGGCAUGAAAACUUAUGUCGGCGGCCUUUGGGCGCAAUUAUUUAUACAUGCCUCAUAAAAAUCGCAAG -------------(((((---((((.---....))).(((((.((.((((((((((....)))))))))).))...))))))))....(((((.........)))))....))).. ( -31.60) >consensus ______UUUUAGUUGCUG___CUGCU___GGUC____GCCCACGCGGCCGGCAUGAAAACUUAUGUCGGCGGCCUUUGGGCGCAAUUAUUUAUACAUGCCUCAUAAAAAUCG_AAA .....................................(((((.((.((((((((((....)))))))))).))...)))))(((............)))................. (-26.55 = -26.05 + -0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:01 2006