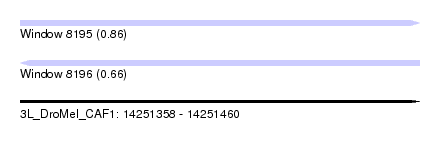
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,251,358 – 14,251,460 |
| Length | 102 |
| Max. P | 0.857699 |

| Location | 14,251,358 – 14,251,460 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 86.08 |
| Mean single sequence MFE | -26.83 |
| Consensus MFE | -22.27 |
| Energy contribution | -23.27 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
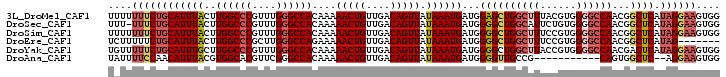
>3L_DroMel_CAF1 14251358 102 + 23771897 CCACUUCCUAUGAGCCGUUGGCCCCACGUAAAGCCAGCUCCAUCAUUUAUAACUGUCAACAGUUUUUGUGGCCCAAACGGGCCAAGUAAAUGCAGAAAAAAA ....(((....((((...((((..........))))))))...((((((((((((....)))))....((((((....)))))).)))))))..)))..... ( -27.70) >DroSec_CAF1 82021 101 + 1 CCACUUCCUAUGAGCCGUUGGCCCCACAGAAUGCCAGCCCCAUCAUUUAUAACUGUCAACAGUUUUUGUGGCCCAAACGGGCCAAGUAAAUGCAGAAA-AAA ....(((.........((((((..........)))))).....((((((((((((....)))))....((((((....)))))).)))))))..))).-... ( -26.70) >DroSim_CAF1 82084 102 + 1 CCACUUCCUAUGAGCCGUUGGCCCCACGGAAAGCCAGCCCCAUCAUUUAUAACUGUCAACAGUUUUUGUGGCCCAAACGGGCCAAGUAAAUGCAGAAAAAAA ....(((.........((((((.(....)...)))))).....((((((((((((....)))))....((((((....)))))).)))))))..)))..... ( -27.00) >DroEre_CAF1 81246 95 + 1 -------CUAUGAGCCGUUGGCCCCACGGAAAGCCAGCCCCAUCAUUUAUAACUGUCAACAGUUUUUCUGGCCCAAGCGGGCCAAGUAAAUGCAGAAAAAGA -------.........((((((.(....)...)))))).....((((((((((((....)))))....((((((....)))))).))))))).......... ( -26.80) >DroYak_CAF1 84748 102 + 1 CCACUUCCUAUGAGUCGUUGGCCCCACGGUAAGCCAGCCCCAUCAUUUAUAACUGUCAACAGUUUUUGUGGCCCAAACGGGCCAAGCAAAUGCAGAAAAACA .........(((((..((((((.(....)...))))))..).))))...............((((((.((((((....)))))).((....))..)))))). ( -26.90) >DroAna_CAF1 78487 89 + 1 CCACUUCCU--GAGCCACUG-----------CGGCAACCCCAUCAUUUAUAACUGUCAACAGUUUUUGUGGCCCGAACGUGCCACGUAAAUGUUGGAAAAUA .........--..(((....-----------.)))....(((.((((((((((((....)))))...(((((.(....).)))))))))))).)))...... ( -25.90) >consensus CCACUUCCUAUGAGCCGUUGGCCCCACGGAAAGCCAGCCCCAUCAUUUAUAACUGUCAACAGUUUUUGUGGCCCAAACGGGCCAAGUAAAUGCAGAAAAAAA ................((((((..........)))))).....((((((((((((....)))))....((((((....)))))).))))))).......... (-22.27 = -23.27 + 1.00)

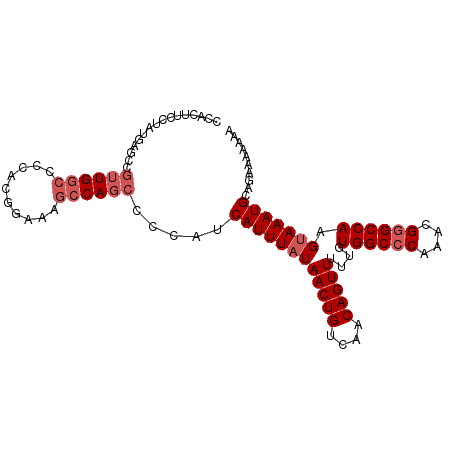
| Location | 14,251,358 – 14,251,460 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 86.08 |
| Mean single sequence MFE | -32.25 |
| Consensus MFE | -26.74 |
| Energy contribution | -28.12 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.655193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14251358 102 - 23771897 UUUUUUUCUGCAUUUACUUGGCCCGUUUGGGCCACAAAAACUGUUGACAGUUAUAAAUGAUGGAGCUGGCUUUACGUGGGGCCAACGGCUCAUAGGAAGUGG ...(((((((((((((..((((((....))))))....(((((....))))).))))))...(((((((((((....))))))...))))).)))))))... ( -34.60) >DroSec_CAF1 82021 101 - 1 UUU-UUUCUGCAUUUACUUGGCCCGUUUGGGCCACAAAAACUGUUGACAGUUAUAAAUGAUGGGGCUGGCAUUCUGUGGGGCCAACGGCUCAUAGGAAGUGG ...-.(((((((((((..((((((....))))))....(((((....))))).)))))).)))))...((.(((((((((.(....).))))))))).)).. ( -32.30) >DroSim_CAF1 82084 102 - 1 UUUUUUUCUGCAUUUACUUGGCCCGUUUGGGCCACAAAAACUGUUGACAGUUAUAAAUGAUGGGGCUGGCUUUCCGUGGGGCCAACGGCUCAUAGGAAGUGG ...((((((.((((((..((((((....))))))....(((((....))))).))))))(((((.(((((((......))))))..).)))))))))))... ( -33.80) >DroEre_CAF1 81246 95 - 1 UCUUUUUCUGCAUUUACUUGGCCCGCUUGGGCCAGAAAAACUGUUGACAGUUAUAAAUGAUGGGGCUGGCUUUCCGUGGGGCCAACGGCUCAUAG------- ..........((((((.(((((((....)))))))...(((((....))))).))))))(((((.(((((((......))))))..).)))))..------- ( -32.10) >DroYak_CAF1 84748 102 - 1 UGUUUUUCUGCAUUUGCUUGGCCCGUUUGGGCCACAAAAACUGUUGACAGUUAUAAAUGAUGGGGCUGGCUUACCGUGGGGCCAACGACUCAUAGGAAGUGG ...((((((.((((((..((((((....))))))....(((((....))))).))))))(((((..((((((......))))))....)))))))))))... ( -32.10) >DroAna_CAF1 78487 89 - 1 UAUUUUCCAACAUUUACGUGGCACGUUCGGGCCACAAAAACUGUUGACAGUUAUAAAUGAUGGGGUUGCCG-----------CAGUGGCUC--AGGAAGUGG ...(((((..((((((.(((((.(....).)))))...(((((....))))).))))))...((((..(..-----------..)..))))--.)))))... ( -28.60) >consensus UUUUUUUCUGCAUUUACUUGGCCCGUUUGGGCCACAAAAACUGUUGACAGUUAUAAAUGAUGGGGCUGGCUUUCCGUGGGGCCAACGGCUCAUAGGAAGUGG ....((((((((((((..((((((....))))))....(((((....))))).))))))...((((((((((......))))))...)))).)))))).... (-26.74 = -28.12 + 1.38)
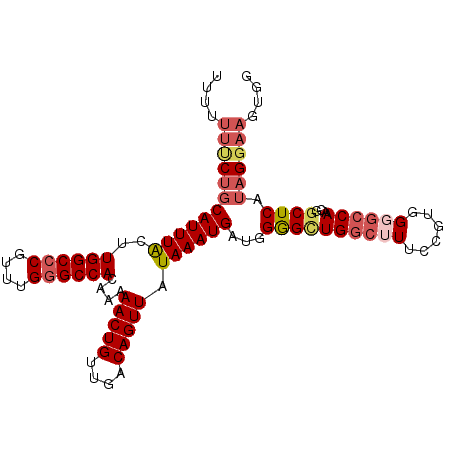
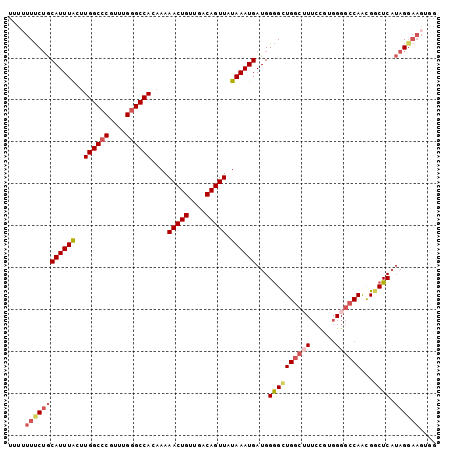
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:50 2006