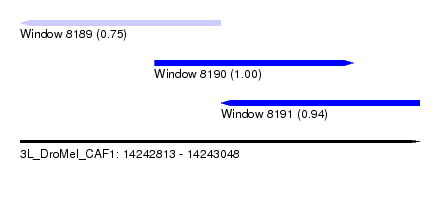
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,242,813 – 14,243,048 |
| Length | 235 |
| Max. P | 0.996902 |

| Location | 14,242,813 – 14,242,931 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 93.04 |
| Mean single sequence MFE | -25.12 |
| Consensus MFE | -19.28 |
| Energy contribution | -20.08 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14242813 118 - 23771897 UUGGCACGAAUUUCAACGAGAGUGACUGGUUAAUUUAGUCAAAAAGGUCUGACUGACAAGCAAAUCGGAAAAUUUCAAAGAGGAAAAUUGAAAAAUUAGCCCAGAUGAAUGGAAUUAA ........(((((((((....))((((((.....))))))......(((((.((((........)))).............((..((((....))))..)))))))...))))))).. ( -21.90) >DroSec_CAF1 73454 117 - 1 UUGCCACGAAU-UCAACGAGAGUGACUGGUUAAUUUAGUCAAAAAGGUCUGGCUGACAAGCCAAUCGGGAAAUAUCAAAGAGGAAAAUUGAAAAAUUAGCCUAGAUGAAUGGAAUUAA ........(((-((..(((...(((((((.....)))))))........(((((....))))).))).......(((...(((..((((....))))..)))...)))...))))).. ( -26.70) >DroSim_CAF1 73535 118 - 1 UUGGCACGAAUUUCAACGAGAGUGACUGGUUAAUUUAGUCAAAAAGGUCUGGCUGACAAGCCAAUCGGGAAAUAUCAAAGAGGAAAAUUGAAAAAUUAGCCUAGAUGAAUGGAAUUAA ........(((((((.(((...(((((((.....)))))))........(((((....))))).))).......(((...(((..((((....))))..)))...))).))))))).. ( -28.10) >DroEre_CAF1 72509 117 - 1 UUUGCACGAAU-UCAACGAGAGUGACUGGUUAAUUUAGUCAAAAAGGUCUGACUGACAAGCCAAUCGGGAAAUUUCAAAGAGGAAAAUUGAAAAAUUAUACUUGAUGACUGAAAUUAA ......(((..-(((.(....)))).(((((...(((((((........)))))))..))))).)))...(((((((..(((...((((....))))...)))......))))))).. ( -21.70) >DroYak_CAF1 75928 118 - 1 UUUGCACGAAUUUCAACAAGAGUGACUGGUUAAUUUAGUCAAAAAGGUCUGACUGACAAGCCAAUCGGGAAACUUCAGAGAUGAAAAUUGAAAAAUUAGCCUUGUUGAAUGGAAUUAA ...........(((((((((.((((.(((((...(((((((........)))))))..))))).)).(....)((((....))))((((....)))).)))))))))))......... ( -27.20) >consensus UUGGCACGAAUUUCAACGAGAGUGACUGGUUAAUUUAGUCAAAAAGGUCUGACUGACAAGCCAAUCGGGAAAUUUCAAAGAGGAAAAUUGAAAAAUUAGCCUAGAUGAAUGGAAUUAA ........(((((((((....))((.(((((...(((((((........)))))))..))))).))........(((...(((..((((....))))..)))...))).))))))).. (-19.28 = -20.08 + 0.80)

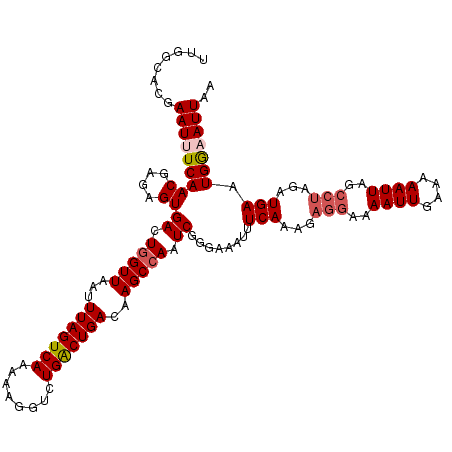
| Location | 14,242,892 – 14,243,009 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 96.75 |
| Mean single sequence MFE | -21.52 |
| Consensus MFE | -19.50 |
| Energy contribution | -19.74 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14242892 117 + 23771897 ACUAAAUUAACCAGUCACUCUCGUUGAAAUUCGUGCCAACAAAAAUCUGGCAUACAAAAUAUGUAAUUUCGCAAAUCAAUUACCCAAACCAAAGGGCAAAACACGGCCGGAUGAAUU ......(((((.((.....)).)))))(((((((....))....(((((((.((((.....)))).................(((........))).........)))))))))))) ( -20.70) >DroSec_CAF1 73533 116 + 1 ACUAAAUUAACCAGUCACUCUCGUUGA-AUUCGUGGCAACAAAAAUCUGGCAUACAAAAUAUGUAAUUUUGCAAAUCAAUUACCCAAACCAAAGGGCAAAACACAGCCGGAUGAAUU ......(((((.((.....)).)))))-((((.((....))...(((((((...((((((.....))))))...........(((........))).........))))))))))). ( -22.80) >DroSim_CAF1 73614 117 + 1 ACUAAAUUAACCAGUCACUCUCGUUGAAAUUCGUGCCAACAAAAAUCUGGCAUACAAAAUAUGUAAUUUCGCAAAUCAAUUACCCAAACCAACGGGCAAAACACAGCCGGAUGAAUU ......(((((.((.....)).)))))(((((((....))....(((((((.((((.....)))).................(((........))).........)))))))))))) ( -20.90) >DroEre_CAF1 72588 116 + 1 ACUAAAUUAACCAGUCACUCUCGUUGA-AUUCGUGCAAACAAAAAUCUGGCAUACAAAAUAUGUAAUUUCGCAAAUCAAUUACCCAAACCAAAGGGCAAAACACGGCCGGAUGAAUU ......(((((.((.....)).)))))-((((((....))....(((((((.((((.....)))).................(((........))).........))))))))))). ( -19.90) >DroYak_CAF1 76007 117 + 1 ACUAAAUUAACCAGUCACUCUUGUUGAAAUUCGUGCAAACAAAAAUCUGGCAUACGAAAUAUGUAAUUUCGCAAAUCAAUUACCCAAACCAAAGGGCAAAACACGGCCGGAUGAAUU ......(((((.((.....)).)))))(((((((....))....(((((((...((((((.....))))))...........(((........))).........)))))))))))) ( -23.30) >consensus ACUAAAUUAACCAGUCACUCUCGUUGAAAUUCGUGCCAACAAAAAUCUGGCAUACAAAAUAUGUAAUUUCGCAAAUCAAUUACCCAAACCAAAGGGCAAAACACGGCCGGAUGAAUU ......(((((.((.....)).)))))(((((((....))....(((((((.((((.....)))).................(((........))).........)))))))))))) (-19.50 = -19.74 + 0.24)


| Location | 14,242,931 – 14,243,048 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 96.76 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -24.64 |
| Energy contribution | -24.52 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14242931 117 - 23771897 GGGAAUUUUCAACUAAUUCCAUUCGGCAUUU-CGCACUCUAAUUCAUCCGGCCGUGUUUUGCCCUUUGGUUUGGGUAAUUGAUUUGCGAAAUUACAUAUUUUGUAUGCCAGAUUUUUG .((((((.......))))))....(((((((-((((...(((((.(((((((((............))).)))))))))))...))))))).((((.....))))))))......... ( -29.60) >DroSec_CAF1 73571 117 - 1 GGGAAUUUUCAACUAAUUCCAUUCGGCAUUU-CUCACUGUAAUUCAUCCGGCUGUGUUUUGCCCUUUGGUUUGGGUAAUUGAUUUGCAAAAUUACAUAUUUUGUAUGCCAGAUUUUUG .((((((.......))))))....(((((..-.....(((((((......((......((((((........)))))).......))..)))))))........)))))......... ( -25.78) >DroSim_CAF1 73653 117 - 1 GGGAAUUUUCAACUAAUUCCAUUCGGCAUUU-CUCACUCUAAUUCAUCCGGCUGUGUUUUGCCCGUUGGUUUGGGUAAUUGAUUUGCGAAAUUACAUAUUUUGUAUGCCAGAUUUUUG .((((((.......))))))....((((...-...((.((((..((.(.(((........))).).))..))))))........((((((((.....))))))))))))......... ( -26.30) >DroEre_CAF1 72626 117 - 1 GGGAAUUUUCAACUAAUUCCAUUCGGCAUUU-CUCACUCUAAUUCAUCCGGCCGUGUUUUGCCCUUUGGUUUGGGUAAUUGAUUUGCGAAAUUACAUAUUUUGUAUGCCAGAUUUUUG .((((((.......))))))....((((...-...((.((((..((...(((........)))...))..))))))........((((((((.....))))))))))))......... ( -24.50) >DroYak_CAF1 76046 118 - 1 GGGAAUUCUCAACUAAUUCCAUUCGGCAUUCACUCACUCUAAUUCAUCCGGCCGUGUUUUGCCCUUUGGUUUGGGUAAUUGAUUUGCGAAAUUACAUAUUUCGUAUGCCAGAUUUUUG .((((((.......))))))....((((.(((.(.((.((((..((...(((........)))...))..)))))).).)))..((((((((.....))))))))))))......... ( -26.70) >consensus GGGAAUUUUCAACUAAUUCCAUUCGGCAUUU_CUCACUCUAAUUCAUCCGGCCGUGUUUUGCCCUUUGGUUUGGGUAAUUGAUUUGCGAAAUUACAUAUUUUGUAUGCCAGAUUUUUG .((((((.......))))))....((((.......((((....(((...(((........)))...)))...))))........((((((((.....))))))))))))......... (-24.64 = -24.52 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:45 2006