| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,527,162 – 1,527,275 |
| Length | 113 |
| Max. P | 0.891607 |

| Location | 1,527,162 – 1,527,275 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.62 |
| Mean single sequence MFE | -32.23 |
| Consensus MFE | -18.32 |
| Energy contribution | -19.02 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
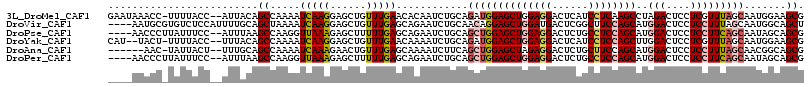
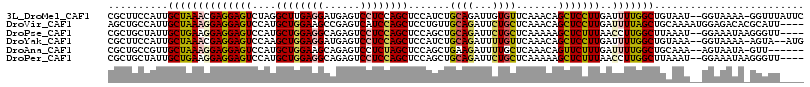
>3L_DroMel_CAF1 1527162 113 + 23771897 GAAUAAACC-UUUUACC--AUUACAGCCAAAAUCAAGGAGCUGUUUGAACACAAUCUGCAGAUGGAGCUGGAGGACUCAUCCUCAAGCCUAGACUCCUCGUUUAGCAAUGGAAGCG .........-.....((--(((...((.(((....(((((((((..((......)).)))).(((.(((.(((((....))))).))))))..)))))..))).)))))))..... ( -27.90) >DroVir_CAF1 8131 112 + 1 ----AAUGCGUGUCUCCAUUUUGCAGCUAAAAUCAAGGAGCUGUUUGAGCAGAAUCUGCAACAGGAGCUGGAUGACUCGGCUUCCAGCAUGGACUCCUCCUUUAGCAAUGGCAGCU ----.......((..(((((.....((((((....((((((((((...((((...)))))))))..((((((.(......).)))))).....)))))..)))))))))))..)). ( -32.50) >DroPse_CAF1 5646 110 + 1 ----AACCCUUAUUUCC--AUUUAAGCCAAGGUUAAAGAGCUUUUUGAGCAGAAUCUGCAGCUGGAGCUGGAGGACUCUGCCUCCAGCAUGGACUCCUCCUUCAGCAAUAGCAGCG ----((((((((.....--...))))....)))).....(((......((((...)))).((((((((((((((......))))))))..(((....)))))))))......))). ( -35.40) >DroYak_CAF1 6190 111 + 1 CAU--UACU-UUUUACC--UUUACAGCCAAAAUCAAGGAGCUGUUUGAACAAAAUCUGCAGAUGGAGCUGGAGGACUCAUCCUCCAGCUUGGACUCCUCGUUUAGCAAUGGAAGCG ...--....-......(--(((...((.(((....(((((((((..((......)).)))).(.(((((((((((....))))))))))).).)))))..))).))....)))).. ( -33.00) >DroAna_CAF1 6418 107 + 1 ------AAC-UAUUACU--UUUGCAGCCAAAAUCAAAGAACUGUUUGAGCAAAAUCUUCAGCUGGAGCUAGAGGACUCUGCUUCCAGCAUGGACUCCUCCUUUAGCAACGGCAGCG ------...-.......--...((.(((....(((((......)))))((..........((((((((.(((....))))).))))))..(((....)))....))...))).)). ( -29.20) >DroPer_CAF1 5816 110 + 1 ----AACCCUUAUUUCC--AUUUAAGCCAAGGUUAAAGAGCUUUUUGAGCAGAAUCUGCAGCUGGAGCUGGAGGACUCUGCCUCCAGCAUGGACUCCUCCUUCAGCAAUAGCAGCG ----((((((((.....--...))))....)))).....(((......((((...)))).((((((((((((((......))))))))..(((....)))))))))......))). ( -35.40) >consensus ____AAACC_UAUUACC__AUUACAGCCAAAAUCAAAGAGCUGUUUGAGCAGAAUCUGCAGCUGGAGCUGGAGGACUCUGCCUCCAGCAUGGACUCCUCCUUUAGCAAUGGCAGCG .........................((.....(((((......)))))............((((((((((((((......))))))))..(((....))))))))).......)). (-18.32 = -19.02 + 0.70)
| Location | 1,527,162 – 1,527,275 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.62 |
| Mean single sequence MFE | -35.03 |
| Consensus MFE | -24.94 |
| Energy contribution | -25.08 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
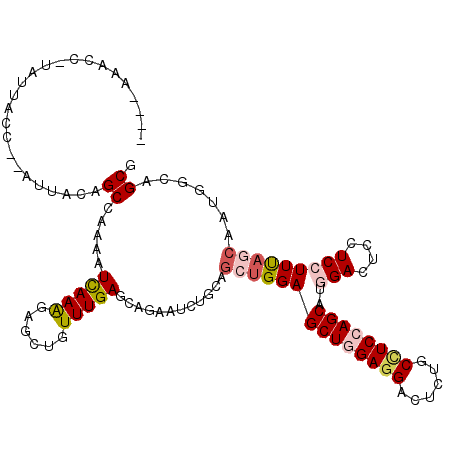
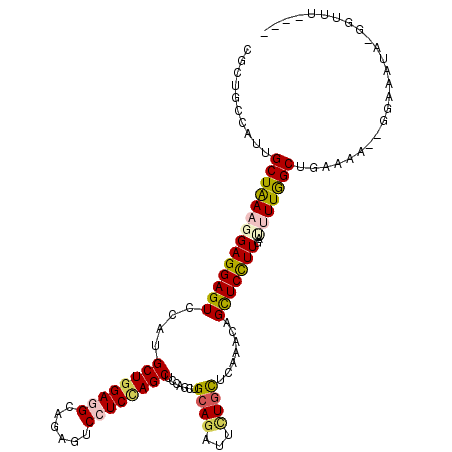
>3L_DroMel_CAF1 1527162 113 - 23771897 CGCUUCCAUUGCUAAACGAGGAGUCUAGGCUUGAGGAUGAGUCCUCCAGCUCCAUCUGCAGAUUGUGUUCAAACAGCUCCUUGAUUUUGGCUGUAAU--GGUAAAA-GGUUUAUUC .(((((((((((((((((((((((...((((.(((((....))))).))))............(((......)))))))))))..))))))...)))--))....)-)))...... ( -32.50) >DroVir_CAF1 8131 112 - 1 AGCUGCCAUUGCUAAAGGAGGAGUCCAUGCUGGAAGCCGAGUCAUCCAGCUCCUGUUGCAGAUUCUGCUCAAACAGCUCCUUGAUUUUAGCUGCAAAAUGGAGACACGCAUU---- .(((((....(((((((((((((.....((((((.(......).))))))..(((((((((...))))...)))))))))))..))))))).)))...((....)).))...---- ( -35.20) >DroPse_CAF1 5646 110 - 1 CGCUGCUAUUGCUGAAGGAGGAGUCCAUGCUGGAGGCAGAGUCCUCCAGCUCCAGCUGCAGAUUCUGCUCAAAAAGCUCUUUAACCUUGGCUUAAAU--GGAAAUAAGGGUU---- ..((((....((((..(((....)))..((((((((......))))))))..)))).)))).....(((.....))).......(((((.((.....--))...)))))...---- ( -35.10) >DroYak_CAF1 6190 111 - 1 CGCUUCCAUUGCUAAACGAGGAGUCCAAGCUGGAGGAUGAGUCCUCCAGCUCCAUCUGCAGAUUUUGUUCAAACAGCUCCUUGAUUUUGGCUGUAAA--GGUAAAA-AGUA--AUG ......(((((((...((((((((...((((((((((....)))))))))).........((......)).....)))))))).(((((.((....)--).)))))-))))--))) ( -39.20) >DroAna_CAF1 6418 107 - 1 CGCUGCCGUUGCUAAAGGAGGAGUCCAUGCUGGAAGCAGAGUCCUCUAGCUCCAGCUGAAGAUUUUGCUCAAACAGUUCUUUGAUUUUGGCUGCAAA--AGUAAUA-GUU------ .(((...((.(((((((..((((.....((((..(((((((((.((.(((....))))).)))))))))....)))).))))..))))))).))...--)))....-...------ ( -33.10) >DroPer_CAF1 5816 110 - 1 CGCUGCUAUUGCUGAAGGAGGAGUCCAUGCUGGAGGCAGAGUCCUCCAGCUCCAGCUGCAGAUUCUGCUCAAAAAGCUCUUUAACCUUGGCUUAAAU--GGAAAUAAGGGUU---- ..((((....((((..(((....)))..((((((((......))))))))..)))).)))).....(((.....))).......(((((.((.....--))...)))))...---- ( -35.10) >consensus CGCUGCCAUUGCUAAAGGAGGAGUCCAUGCUGGAGGCAGAGUCCUCCAGCUCCAGCUGCAGAUUCUGCUCAAACAGCUCCUUGAUUUUGGCUGAAAA__GGAAAUA_GGUUU____ ..........((((((((((((((....((((((((......)))))))).......((((...)))).......)))))))..)))))))......................... (-24.94 = -25.08 + 0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:02 2006