| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,212,630 – 14,212,725 |
| Length | 95 |
| Max. P | 0.993355 |

| Location | 14,212,630 – 14,212,725 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 73.13 |
| Mean single sequence MFE | -31.47 |
| Consensus MFE | -13.70 |
| Energy contribution | -13.87 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.44 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
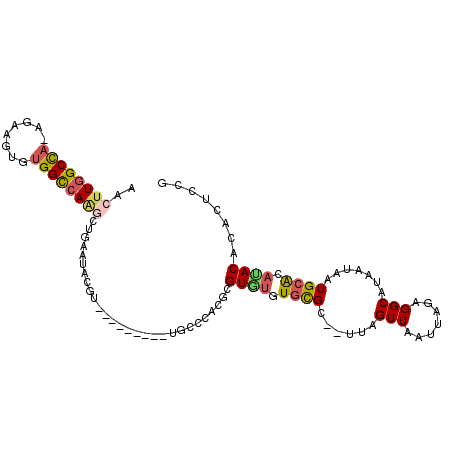
>3L_DroMel_CAF1 14212630 95 + 23771897 AACUUGGCCA-AGAAGUGUGGCCAAGCUGAGUAUGU--------CUGUCUAUGCGUGUGUGCGC--UUAGUUAAUUAGCGGCAUAAUAAUGCACACACACACUCCG ..((((((((-.......))))))))....(((((.--------.....)))))((((((((((--(.........))))((((....)))).)))))))...... ( -33.10) >DroPse_CAF1 46329 98 + 1 AACUUGGCCAAAGAAGUGUGGCCAACGCAAAUGCUCAUA------CACGCACGUGUAUGUGCGU--CUAGUUAAUUACAGGCAUAAUAAUGCGGAUACACUCUCCU ...(((((((........)))))))((((...((.((((------(((....))))))).))((--((.((.....)))))).......))))............. ( -32.20) >DroSec_CAF1 47773 95 + 1 AACUUGGCCA-AGAAGUGUGGCCAAGCUGAGGAUGU--------AUGCCCAUGCGUGUGUGCGC--UUAGUUAAUUAGCGGCGUAAUAAUGCACACACACACUCGG ..((((((((-.......))))))))(((((...((--------((....))))((((((((((--(.........))))((((....)))).))))))).))))) ( -37.60) >DroEre_CAF1 46834 104 + 1 AACUUGGCUA-AGAAGUGUGGCCAGGAUGAGUACGUAUGUAUGUGUGUCCAUGCGUGUGUGUGCUC-UAGUUAAUUAGCGGCGUAAUAAUGCACAUACACACUCCG ..((((((((-.......))))))))..((((..(((((.((....)).)))))((((((((((..-..((((..........))))...)))))))))))))).. ( -33.50) >DroAna_CAF1 49051 94 + 1 AACUUGGCCAAAGAAGUGCGGUGAUGCUGAACA--A--------AAGCACACACGUUGAUAUGC--UUAGUUAAUUAGAGGCAUAAUAAUGCAGAUACACUCUCGC ............((((((..(((.((((.....--.--------.))))))).(((((.(((((--((..........))))))).)))))......)))).)).. ( -20.20) >DroPer_CAF1 54903 98 + 1 AACUUGGCCAAAGAAGUGUGGCCAACGCAAAUGCUCAUA------CACGCACGUGUAUGUGCGU--CUAGUUAAUUACAGGCAUAAUAAUGCGGAUACACUCUCCU ...(((((((........)))))))((((...((.((((------(((....))))))).))((--((.((.....)))))).......))))............. ( -32.20) >consensus AACUUGGCCA_AGAAGUGUGGCCAAGCUGAAUACGU_________UGCCCACGCGUGUGUGCGC__UUAGUUAAUUAGAGGCAUAAUAAUGCACAUACACACUCCG ..((((((((........))))))))............................((((.((((......(((.......))).......)))).))))........ (-13.70 = -13.87 + 0.17)


| Location | 14,212,630 – 14,212,725 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 73.13 |
| Mean single sequence MFE | -29.05 |
| Consensus MFE | -10.97 |
| Energy contribution | -12.44 |
| Covariance contribution | 1.47 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.38 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.946982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
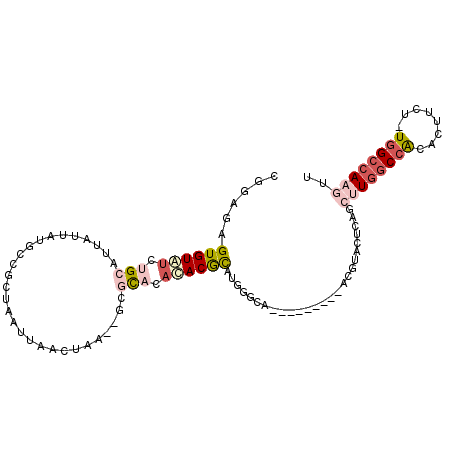
>3L_DroMel_CAF1 14212630 95 - 23771897 CGGAGUGUGUGUGUGCAUUAUUAUGCCGCUAAUUAACUAA--GCGCACACACGCAUAGACAG--------ACAUACUCAGCUUGGCCACACUUCU-UGGCCAAGUU ..((((((((.(((((((....))))..........(((.--(((......))).)))))).--------))))))))((((((((((.......-)))))))))) ( -38.20) >DroPse_CAF1 46329 98 - 1 AGGAGAGUGUAUCCGCAUUAUUAUGCCUGUAAUUAACUAG--ACGCACAUACACGUGCGUG------UAUGAGCAUUUGCGUUGGCCACACUUCUUUGGCCAAGUU .(((.......)))((((....))))........((((.(--((((((((((((....)))------))))......))))))(((((........))))).)))) ( -29.30) >DroSec_CAF1 47773 95 - 1 CCGAGUGUGUGUGUGCAUUAUUACGCCGCUAAUUAACUAA--GCGCACACACGCAUGGGCAU--------ACAUCCUCAGCUUGGCCACACUUCU-UGGCCAAGUU (((.((((((((((((........................--)))))))))))).)))....--------........((((((((((.......-)))))))))) ( -37.46) >DroEre_CAF1 46834 104 - 1 CGGAGUGUGUAUGUGCAUUAUUACGCCGCUAAUUAACUA-GAGCACACACACGCAUGGACACACAUACAUACGUACUCAUCCUGGCCACACUUCU-UAGCCAAGUU .(((((((((.(((((.(((.................))-).))))).))))))(((......))).............)))((((.........-..)))).... ( -21.93) >DroAna_CAF1 49051 94 - 1 GCGAGAGUGUAUCUGCAUUAUUAUGCCUCUAAUUAACUAA--GCAUAUCAACGUGUGUGCUU--------U--UGUUCAGCAUCACCGCACUUCUUUGGCCAAGUU ((((((((((....((((....))))........(((.((--((((((......))))))))--------.--.)))..........)))).))))..))...... ( -18.10) >DroPer_CAF1 54903 98 - 1 AGGAGAGUGUAUCCGCAUUAUUAUGCCUGUAAUUAACUAG--ACGCACAUACACGUGCGUG------UAUGAGCAUUUGCGUUGGCCACACUUCUUUGGCCAAGUU .(((.......)))((((....))))........((((.(--((((((((((((....)))------))))......))))))(((((........))))).)))) ( -29.30) >consensus CGGAGAGUGUAUCUGCAUUAUUAUGCCGCUAAUUAACUAA__GCGCACACACGCAUGGGCA_________ACGUACUCAGCUUGGCCACACUUCU_UGGCCAAGUU ......((((((.(((............................))).))))))..........................((((((((........)))))))).. (-10.97 = -12.44 + 1.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:37 2006