
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,199,074 – 14,199,348 |
| Length | 274 |
| Max. P | 0.997762 |

| Location | 14,199,074 – 14,199,178 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 87.12 |
| Mean single sequence MFE | -30.27 |
| Consensus MFE | -20.08 |
| Energy contribution | -20.11 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.49 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14199074 104 + 23771897 UCGGAUUCUGGUAUCCCUAACCAUGGAAUCCCAAAUCCUGGCAUCCCUAACCAUGGAAUACCGAAUCGUGGAAUUCCUAAUGAUGGAAUCCCAAAUCCUGGCAU ...(((((.(((((.((.......((.((.(((.....))).)).)).......)).)))))))))).(((.(((((.......))))).)))........... ( -26.84) >DroPse_CAF1 33114 104 + 1 CCAAAUCCUGGUAUUCCCAACGAAGGAAUCCCAAAUCCUGGCAUUCCCAAUAGGGGAAUACCAAAUCCUGGCAUUCCCAACGAAGGAAUCCCAAAUCCUGGCAU (((.....((((((((((......(((((.(((.....))).)))))......)))))))))).....(((.(((((.......))))).))).....)))... ( -34.70) >DroSim_CAF1 33252 104 + 1 UCGGAUUCUGGUAUUCCUAACCAUGGAAUCCCAAAUCCUGGCAUCCCUAACCAUGGAAUACCGAAUCGUGGAAUUCCUAAUGAUGGAAUCCCAAAUCCUGGCAU ...(((((.((((((((.......((.((.(((.....))).)).)).......))))))))))))).(((.(((((.......))))).)))........... ( -31.24) >DroEre_CAF1 33989 104 + 1 UCGGAUUCUGGUAUCCCUAACCAUGGCAUCCCAAAUCCUGGCAUCCCUAACCAUGGAAUACCGAAUCGUGGAAUCCCCAAUGAUGGAAUCCCAAAUCCUGGCAU ..((((((((((((.((.......((.((.(((.....))).)).)).......)).)))))..(((((((.....)).))))))))))))............. ( -26.24) >DroYak_CAF1 35825 104 + 1 UCGGAUUCUGGUAUCCCUAACAGUGGAAUCCCAAAUCCUGGCAUCCCCAACCAUGGAAUACCGAAUCGUGGAAUCCCCAACGAUGGAAUCCCGAAUCCUGGAAU ((((((((((((((.((......(((.((.(((.....))).))..))).....)).)))))..(((((((.....)).)))))))))).)))).......... ( -28.40) >DroPer_CAF1 41729 104 + 1 CCAAAUCCUGGCAUUCCCAACGAAGGAAUCCCAAAUCCUGGUAUCCCCAAUAGGGGAAUACCAAAUCCUGGCAUUCCCAACGAAGGAAUCCCAAAUCCUGGCAU (((.....(((.(((((...((..(((((.(((.....((((((((((....)))).)))))).....))).)))))...))..))))).))).....)))... ( -34.20) >consensus UCGGAUUCUGGUAUCCCUAACCAUGGAAUCCCAAAUCCUGGCAUCCCCAACCAUGGAAUACCGAAUCGUGGAAUUCCCAACGAUGGAAUCCCAAAUCCUGGCAU ((((....((((((.((.......((.((.(((.....))).)).)).......)).)))))).....(((.(((((.......))))).)))....))))... (-20.08 = -20.11 + 0.03)


| Location | 14,199,074 – 14,199,178 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 87.12 |
| Mean single sequence MFE | -46.47 |
| Consensus MFE | -40.62 |
| Energy contribution | -39.46 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.26 |
| Mean z-score | -5.52 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.997006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14199074 104 - 23771897 AUGCCAGGAUUUGGGAUUCCAUCAUUAGGAAUUCCACGAUUCGGUAUUCCAUGGUUAGGGAUGCCAGGAUUUGGGAUUCCAUGGUUAGGGAUACCAGAAUCCGA ......((((((.((((((((((((..(((((((((.(((((((((((((.......)))))))).)))))))))))))))))))..))))).)).)))))).. ( -44.50) >DroPse_CAF1 33114 104 - 1 AUGCCAGGAUUUGGGAUUCCUUCGUUGGGAAUGCCAGGAUUUGGUAUUCCCCUAUUGGGAAUGCCAGGAUUUGGGAUUCCUUCGUUGGGAAUACCAGGAUUUGG ...(((((.(((((.((((((.((..((((((.(((((.((((((((((((.....)))))))))))).))))).)))))).))..)))))).))))).))))) ( -50.40) >DroSim_CAF1 33252 104 - 1 AUGCCAGGAUUUGGGAUUCCAUCAUUAGGAAUUCCACGAUUCGGUAUUCCAUGGUUAGGGAUGCCAGGAUUUGGGAUUCCAUGGUUAGGAAUACCAGAAUCCGA ......((((((.((((((((((((..(((((((((.(((((((((((((.......)))))))).)))))))))))))))))))..))))).)).)))))).. ( -45.00) >DroEre_CAF1 33989 104 - 1 AUGCCAGGAUUUGGGAUUCCAUCAUUGGGGAUUCCACGAUUCGGUAUUCCAUGGUUAGGGAUGCCAGGAUUUGGGAUGCCAUGGUUAGGGAUACCAGAAUCCGA ......((((((.((((((((((((..((.((((((.(((((((((((((.......)))))))).))))))))))).)))))))..))))).)).)))))).. ( -42.20) >DroYak_CAF1 35825 104 - 1 AUUCCAGGAUUCGGGAUUCCAUCGUUGGGGAUUCCACGAUUCGGUAUUCCAUGGUUGGGGAUGCCAGGAUUUGGGAUUCCACUGUUAGGGAUACCAGAAUCCGA ......((((((.(((((((.......(((((((((.(((((((((((((.......)))))))).)))))))))))))).......))))).)).)))))).. ( -44.44) >DroPer_CAF1 41729 104 - 1 AUGCCAGGAUUUGGGAUUCCUUCGUUGGGAAUGCCAGGAUUUGGUAUUCCCCUAUUGGGGAUACCAGGAUUUGGGAUUCCUUCGUUGGGAAUGCCAGGAUUUGG ...(((((.(((((.((((((.((..((((((.(((((.((((((((.((((....)))))))))))).))))).)))))).))..)))))).))))).))))) ( -52.30) >consensus AUGCCAGGAUUUGGGAUUCCAUCAUUGGGAAUUCCACGAUUCGGUAUUCCAUGGUUAGGGAUGCCAGGAUUUGGGAUUCCAUGGUUAGGAAUACCAGAAUCCGA ......((((((.(((((((.......(((((((((.(((((((((((((.......)))))))).)))))))))))))).......))))).)).)))))).. (-40.62 = -39.46 + -1.16)

| Location | 14,199,113 – 14,199,218 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 78.60 |
| Mean single sequence MFE | -26.76 |
| Consensus MFE | -13.70 |
| Energy contribution | -14.03 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14199113 105 + 23771897 GGCAUCCCUAACCAUGGAAUACCGAAUCGUGGAAUUCCUAAUGAUGGAAUCCCAAAUCCUGGCAUUCCUAAUCAUGGAAUUCCAAAUCCUGGAAUCCCUAAUCAU ((..(((........)))...(((.....(((((((((..((((((((((.(((.....))).)))))..)))))))))))))).....)))....))....... ( -29.00) >DroPse_CAF1 33153 105 + 1 GGCAUUCCCAAUAGGGGAAUACCAAAUCCUGGCAUUCCCAACGAAGGAAUCCCAAAUCCUGGCAUUCCCAACGAAGGAAUCCCAAAUCCUGGUAUCCCCAAUAGG .......((.((.(((((.(((((.....(((.(((((...((..(((((.(((.....))).)))))...))..))))).))).....)))))))))).)).)) ( -33.80) >DroEre_CAF1 34028 105 + 1 GGCAUCCCUAACCAUGGAAUACCGAAUCGUGGAAUCCCCAAUGAUGGAAUCCCAAAUCCUGGCAUUCCCAAUCAUGGAAUUCCCAAUCCUGGAAUCCCUAAUCAU ((.((.((.......(((((.((......(((.....)))((((((((((.(((.....))).)))))..))))))).))))).......)).)).))....... ( -24.14) >DroWil_CAF1 43859 101 + 1 --GAU--ACAAUCACGGUAUUCCCAAUCCAGGCAUCCCCAAUCAAGGUAUUCCCAAUCCAGGCAUACCAAACCGUGGUCUACCCAAUCCUGGCAUCCCCAACGAA --(((--...((((((((............((.....))......(((((.((.......)).)))))..))))))))....(((....))).)))......... ( -19.50) >DroYak_CAF1 35864 105 + 1 GGCAUCCCCAACCAUGGAAUACCGAAUCGUGGAAUCCCCAACGAUGGAAUCCCGAAUCCUGGAAUCCCCAAUCAUGGAAUUCCCAAUCCUGGAAUCCCUAAUCAU ((.....))..(((.(((.....((.(((.(((.(((........))).)))))).))..(((((..(((....))).)))))...))))))............. ( -24.20) >DroPer_CAF1 41768 105 + 1 GGUAUCCCCAAUAGGGGAAUACCAAAUCCUGGCAUUCCCAACGAAGGAAUCCCAAAUCCUGGCAUCCCCAAUGUAGGAAUACCGAAUCCUGGCAUUCCCAACGAA (((((((((....)))).)))))......(((.(((((.......))))).))).....(((.....))).(((.(((((.(((.....))).)))))..))).. ( -29.90) >consensus GGCAUCCCCAACCAUGGAAUACCGAAUCCUGGAAUCCCCAACGAAGGAAUCCCAAAUCCUGGCAUUCCCAAUCAUGGAAUUCCCAAUCCUGGAAUCCCCAAUCAU ...............(((((.((......(((.....))).....(((((.(((.....))).))))).......)).)))))......(((.....)))..... (-13.70 = -14.03 + 0.33)

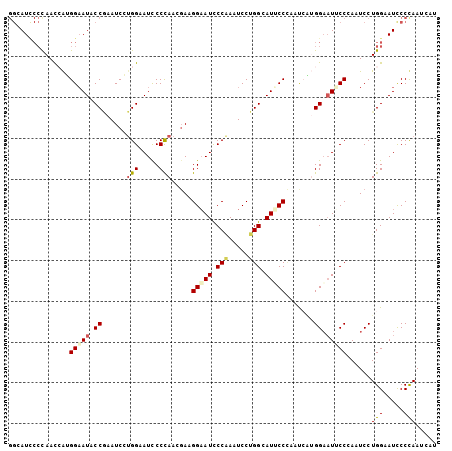
| Location | 14,199,113 – 14,199,218 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 78.60 |
| Mean single sequence MFE | -43.08 |
| Consensus MFE | -31.72 |
| Energy contribution | -31.58 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.37 |
| Mean z-score | -4.53 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.975005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14199113 105 - 23771897 AUGAUUAGGGAUUCCAGGAUUUGGAAUUCCAUGAUUAGGAAUGCCAGGAUUUGGGAUUCCAUCAUUAGGAAUUCCACGAUUCGGUAUUCCAUGGUUAGGGAUGCC .((((((((((((((((...)))))))))).))))))((((((((.((((((((((((((.......))))))))).)))))))))))))..(((.......))) ( -44.10) >DroPse_CAF1 33153 105 - 1 CCUAUUGGGGAUACCAGGAUUUGGGAUUCCUUCGUUGGGAAUGCCAGGAUUUGGGAUUCCUUCGUUGGGAAUGCCAGGAUUUGGUAUUCCCCUAUUGGGAAUGCC ((((..((((((((((((.(((((.((((((.((..((((((.((((...)))).)))))).))..)))))).))))).))))))).)))))...))))...... ( -50.10) >DroEre_CAF1 34028 105 - 1 AUGAUUAGGGAUUCCAGGAUUGGGAAUUCCAUGAUUGGGAAUGCCAGGAUUUGGGAUUCCAUCAUUGGGGAUUCCACGAUUCGGUAUUCCAUGGUUAGGGAUGCC ..(((((((((((((.......)))))))).))))).((((((((.((((((((((((((.......))))))))).)))))))))))))..(((.......))) ( -41.10) >DroWil_CAF1 43859 101 - 1 UUCGUUGGGGAUGCCAGGAUUGGGUAGACCACGGUUUGGUAUGCCUGGAUUGGGAAUACCUUGAUUGGGGAUGCCUGGAUUGGGAAUACCGUGAUUGU--AUC-- .......((..((((.......))))..))((((((((((((.((..(.(..((....(((.....)))....))..).)..)).)))))).))))))--...-- ( -33.00) >DroYak_CAF1 35864 105 - 1 AUGAUUAGGGAUUCCAGGAUUGGGAAUUCCAUGAUUGGGGAUUCCAGGAUUCGGGAUUCCAUCGUUGGGGAUUCCACGAUUCGGUAUUCCAUGGUUGGGGAUGCC .......((((((((.(((((.((((((((.......))))))))..)))))))))))))(((((.((.....)))))))..((((((((.......)))))))) ( -41.70) >DroPer_CAF1 41768 105 - 1 UUCGUUGGGAAUGCCAGGAUUCGGUAUUCCUACAUUGGGGAUGCCAGGAUUUGGGAUUCCUUCGUUGGGAAUGCCAGGAUUUGGUAUUCCCCUAUUGGGGAUACC .((.((((.....))))))...((((((((((....((((((((((((.(((((.((((((.....)))))).))))).))))))))))))....)))))))))) ( -48.50) >consensus AUGAUUAGGGAUUCCAGGAUUGGGAAUUCCAUGAUUGGGAAUGCCAGGAUUUGGGAUUCCAUCAUUGGGAAUGCCACGAUUCGGUAUUCCAUGAUUGGGGAUGCC .......((((((((.......)))))))).((((((((((((((.((((((((.(((((.......))))).))).))))))))))))).))))))........ (-31.72 = -31.58 + -0.13)


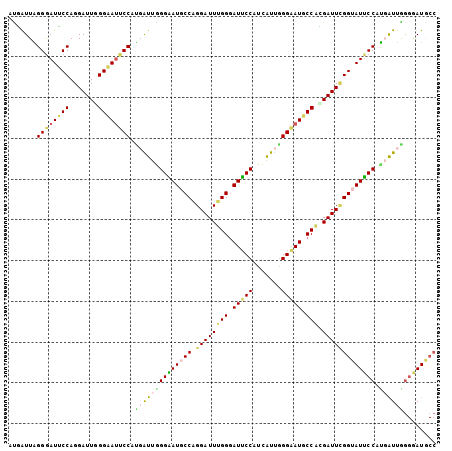
| Location | 14,199,138 – 14,199,258 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.00 |
| Mean single sequence MFE | -33.87 |
| Consensus MFE | -11.83 |
| Energy contribution | -12.06 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.53 |
| Structure conservation index | 0.35 |
| SVM decision value | 2.93 |
| SVM RNA-class probability | 0.997762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14199138 120 + 23771897 AUCGUGGAAUUCCUAAUGAUGGAAUCCCAAAUCCUGGCAUUCCUAAUCAUGGAAUUCCAAAUCCUGGAAUCCCUAAUCAUGGAAUUCCAAACCGUGGCAUUCCUAAUGAUGGAAUCCCGA .((((((((((((..((((((((((.(((.....))).)))))..))))))))))))))......(((.(((...((((((((((.(((.....))).)))))..)))))))).)))))) ( -41.70) >DroPse_CAF1 33178 120 + 1 AUCCUGGCAUUCCCAACGAAGGAAUCCCAAAUCCUGGCAUUCCCAACGAAGGAAUCCCAAAUCCUGGUAUCCCCAAUAGGGGAAUACCAAAUCCUGGCAUUCCCAACGAAGGAAUCCCAA ....(((.(((((...((..(((((.(((.....))).))))).......(((((.(((.....((((((((((....)))).)))))).....))).)))))...))..))))).))). ( -40.90) >DroYak_CAF1 35889 120 + 1 AUCGUGGAAUCCCCAACGAUGGAAUCCCGAAUCCUGGAAUCCCCAAUCAUGGAAUUCCCAAUCCUGGAAUCCCUAAUCAUGGAAUUCCCAACCGUGGCAUUCCUAAUGAUGGAAUCCCGA .(((.(((.(((........))).))))))...(.(((.(((...((((((((((..(((.....(((((.((.......)).)))))......))).)))))..)))))))).))).). ( -33.90) >DroMoj_CAF1 44328 105 + 1 ACCCGGGCAUUCCUAAUCCCGGUAUGCCCAAUCCUGGCAUUCCAAACCGCGGCAUGCCCAAUCCUGGAAUCGAAAAUCCCGGAAUGCCCAAUCCUGGUAUCCCGA--------------- ....(((((((((.......((((((((......(((....)))......)))))))).......(((........))).))))))))).....(((....))).--------------- ( -35.10) >DroAna_CAF1 31368 120 + 1 AUCCAGGCAUCCCCAAUCAUGGUAUUCCGAAUCGUGGCAUUCCCAAUGACGGCAUUCCUAACCCUGGAAUUCCCAUUGACGGUAUUCCCAAUCCUGGUAUCCCCAAUCAUGGAAUUCCCA ..(((((.........(((((((.......)))))))......(((((..((.(((((.......))))).)))))))..((.....))...))))).....(((....)))........ ( -24.20) >DroPer_CAF1 41793 120 + 1 AUCCUGGCAUUCCCAACGAAGGAAUCCCAAAUCCUGGCAUCCCCAAUGUAGGAAUACCGAAUCCUGGCAUUCCCAACGAAGGAAUCCCAAAUCCUGGCAUCCCCAAUGUAGGAAUACCGA ....(((.(((((.......))))).)))..(((((.(((.....))))))))....((.....(((.(((((.......))))).)))..(((((.(((.....))))))))....)). ( -27.40) >consensus AUCCUGGCAUUCCCAACGAUGGAAUCCCAAAUCCUGGCAUUCCCAAUCAAGGAAUUCCAAAUCCUGGAAUCCCCAAUCACGGAAUUCCAAAUCCUGGCAUCCCCAAUGAUGGAAUCCCGA ....(((.((.((.......((.((.(((.....))).)).)).......)).)).)))......(((((.((.......)).)))))................................ (-11.83 = -12.06 + 0.22)


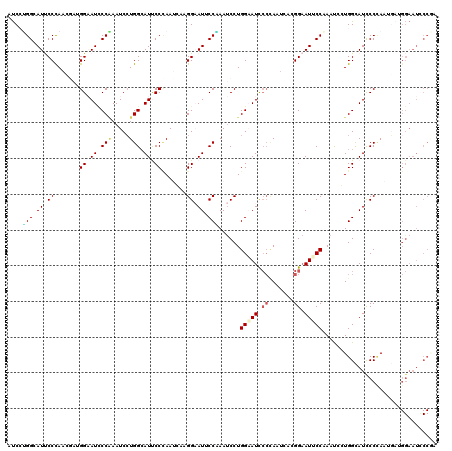
| Location | 14,199,138 – 14,199,258 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.00 |
| Mean single sequence MFE | -49.76 |
| Consensus MFE | -31.86 |
| Energy contribution | -34.12 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.27 |
| Mean z-score | -4.70 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14199138 120 - 23771897 UCGGGAUUCCAUCAUUAGGAAUGCCACGGUUUGGAAUUCCAUGAUUAGGGAUUCCAGGAUUUGGAAUUCCAUGAUUAGGAAUGCCAGGAUUUGGGAUUCCAUCAUUAGGAAUUCCACGAU ..((((((((.......(((((.(((...(((((.(((((.((((((((((((((((...)))))))))).))))))))))).)))))...))).))))).......))))))))..... ( -50.44) >DroPse_CAF1 33178 120 - 1 UUGGGAUUCCUUCGUUGGGAAUGCCAGGAUUUGGUAUUCCCCUAUUGGGGAUACCAGGAUUUGGGAUUCCUUCGUUGGGAAUGCCAGGAUUUGGGAUUCCUUCGUUGGGAAUGCCAGGAU ((((.((((((.((..((((((.(((((.((((((((.((((....)))))))))))).))))).)))))).))..)))))).))))..(((((.((((((.....)))))).))))).. ( -60.20) >DroYak_CAF1 35889 120 - 1 UCGGGAUUCCAUCAUUAGGAAUGCCACGGUUGGGAAUUCCAUGAUUAGGGAUUCCAGGAUUGGGAAUUCCAUGAUUGGGGAUUCCAGGAUUCGGGAUUCCAUCGUUGGGGAUUCCACGAU ..((.(((((.......))))).))...(((((((((((((((((..((((((((.(((((.((((((((.......))))))))..))))))))))))))))))..)))))))).)))) ( -51.40) >DroMoj_CAF1 44328 105 - 1 ---------------UCGGGAUACCAGGAUUGGGCAUUCCGGGAUUUUCGAUUCCAGGAUUGGGCAUGCCGCGGUUUGGAAUGCCAGGAUUGGGCAUACCGGGAUUAGGAAUGCCCGGGU ---------------..((....))....(((((((((((((((((...))))))((..((((..(((((.((((((((....))).)))))))))).))))..)).))))))))))).. ( -42.50) >DroAna_CAF1 31368 120 - 1 UGGGAAUUCCAUGAUUGGGGAUACCAGGAUUGGGAAUACCGUCAAUGGGAAUUCCAGGGUUAGGAAUGCCGUCAUUGGGAAUGCCACGAUUCGGAAUACCAUGAUUGGGGAUGCCUGGAU ..((.(((((.....(((.....)))((((((((....(((...((((..(((((.......))))).))))...))).....)).))))))))))).))....(..((....))..).. ( -35.30) >DroPer_CAF1 41793 120 - 1 UCGGUAUUCCUACAUUGGGGAUGCCAGGAUUUGGGAUUCCUUCGUUGGGAAUGCCAGGAUUCGGUAUUCCUACAUUGGGGAUGCCAGGAUUUGGGAUUCCUUCGUUGGGAAUGCCAGGAU ..((((((((((....((((((.(((((.(((((.((((((....((((((((((.......))))))))))....)))))).))))).))))).))))))....))))))))))..... ( -58.70) >consensus UCGGGAUUCCAUCAUUGGGAAUGCCAGGAUUGGGAAUUCCAUGAUUGGGGAUUCCAGGAUUGGGAAUUCCAUCAUUGGGAAUGCCAGGAUUUGGGAUUCCAUCAUUGGGAAUGCCAGGAU ..((.(((((.......))))).))....(((((((((((.......((((((((.......)))))))).......(((((.((.......)).))))).......))))))))))).. (-31.86 = -34.12 + 2.25)

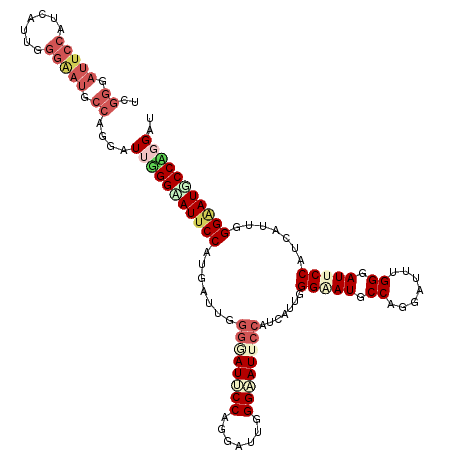
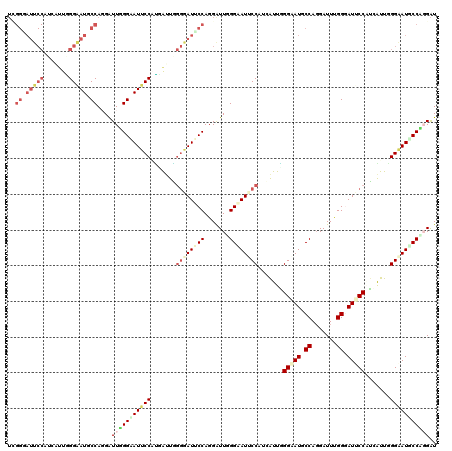
| Location | 14,199,178 – 14,199,298 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.72 |
| Mean single sequence MFE | -31.56 |
| Consensus MFE | -21.75 |
| Energy contribution | -22.89 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961376 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14199178 120 + 23771897 UCCUAAUCAUGGAAUUCCAAAUCCUGGAAUCCCUAAUCAUGGAAUUCCAAACCGUGGCAUUCCUAAUGAUGGAAUCCCGAAUCCUGGCAUUCCCAAUCACGGUAUUCCCAACCCUGGAAU (((......((((((((((......((....))......)))))))))).((((((((((.....)))..(((((.(((.....))).)))))...)))))))............))).. ( -34.30) >DroPse_CAF1 33218 120 + 1 UCCCAACGAAGGAAUCCCAAAUCCUGGUAUCCCCAAUAGGGGAAUACCAAAUCCUGGCAUUCCCAACGAAGGAAUCCCAAAUCCUGGCAUCCCCAAUGAAGGAAUACCGAAUCCUGGCAU ..((..((..(((((.(((.....((((((((((....)))).)))))).....))).)))))...)).((((.((.....((((..(((.....))).)))).....)).))))))... ( -35.50) >DroSec_CAF1 32926 120 + 1 UCCUAAUCAUGGAAUUCCCAAUCCUGGAAUCCCUAAUCAUGGAAUUCCAAACCGUGGCAUUCCUAAUGAUGGAAUCCCUAAUCCUGGCAUUCCUAAUCAUGGAAUUCCAAACCGUGGCAU ......((((((............((((((.((.......)).)))))).....(((.(((((..((((((((((.((.......)).)))))..)))))))))).)))..))))))... ( -34.10) >DroYak_CAF1 35929 120 + 1 CCCCAAUCAUGGAAUUCCCAAUCCUGGAAUCCCUAAUCAUGGAAUUCCCAACCGUGGCAUUCCUAAUGAUGGAAUCCCGAAUCCUGGCAUUCCCAACCAUGGAAUUCCCAAUCCUGGAAU ..(((.....((((((((.......(((((.((......(((.....)))..((.((.(((((.......)))))))))......)).))))).......))))))))......)))... ( -31.84) >DroAna_CAF1 31408 120 + 1 UCCCAAUGACGGCAUUCCUAACCCUGGAAUUCCCAUUGACGGUAUUCCCAAUCCUGGUAUCCCCAAUCAUGGAAUUCCCAAUCCUGGUAUCCCCAAUCAUGGCAUUCCGAAUCGUGGCAU ...(((((..((.(((((.......))))).)))))))(((((....(((....)))............((((((..(((....(((.....)))....))).)))))).)))))..... ( -25.50) >DroPer_CAF1 41833 120 + 1 CCCCAAUGUAGGAAUACCGAAUCCUGGCAUUCCCAACGAAGGAAUCCCAAAUCCUGGCAUCCCCAAUGUAGGAAUACCGAAUCCUGGCAUUCCCAACGAAGGAAUCCCAAAUCCUGGCAU .....((((((((.....((.((.(((.(((((.......))))).)))..(((((.(((.....)))))))).....)).)).(((.(((((.......))))).)))..)))).)))) ( -28.10) >consensus UCCCAAUCAUGGAAUUCCAAAUCCUGGAAUCCCCAAUCAUGGAAUUCCAAACCCUGGCAUUCCCAAUGAUGGAAUCCCGAAUCCUGGCAUUCCCAAUCAUGGAAUUCCAAAUCCUGGCAU ..(((.....(((........))).(((((.((.......(((((.(((.....(((.(((((.......))))).))).....))).))))).......)).)))))......)))... (-21.75 = -22.89 + 1.14)

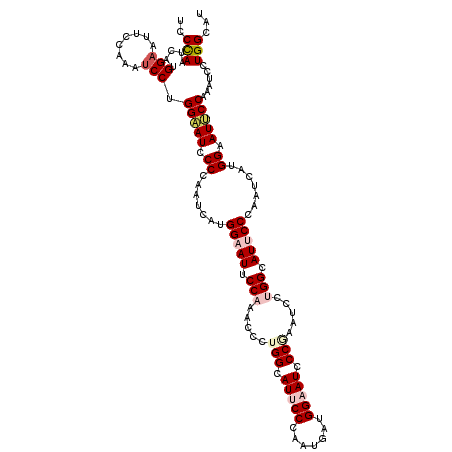

| Location | 14,199,178 – 14,199,298 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.72 |
| Mean single sequence MFE | -46.48 |
| Consensus MFE | -33.77 |
| Energy contribution | -34.30 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.31 |
| Mean z-score | -4.06 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.970204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14199178 120 - 23771897 AUUCCAGGGUUGGGAAUACCGUGAUUGGGAAUGCCAGGAUUCGGGAUUCCAUCAUUAGGAAUGCCACGGUUUGGAAUUCCAUGAUUAGGGAUUCCAGGAUUUGGAAUUCCAUGAUUAGGA ((((((((....(((((.((.(((((((((((.(((((....((.(((((.......))))).))....))))).))))).)))))).)))))))....))))))))(((.......))) ( -42.80) >DroPse_CAF1 33218 120 - 1 AUGCCAGGAUUCGGUAUUCCUUCAUUGGGGAUGCCAGGAUUUGGGAUUCCUUCGUUGGGAAUGCCAGGAUUUGGUAUUCCCCUAUUGGGGAUACCAGGAUUUGGGAUUCCUUCGUUGGGA ...((..((((((((((((((.....))))))))).)))))..))..((((.((..((((((.(((((.((((((((.((((....)))))))))))).))))).)))))).))..)))) ( -57.20) >DroSec_CAF1 32926 120 - 1 AUGCCACGGUUUGGAAUUCCAUGAUUAGGAAUGCCAGGAUUAGGGAUUCCAUCAUUAGGAAUGCCACGGUUUGGAAUUCCAUGAUUAGGGAUUCCAGGAUUGGGAAUUCCAUGAUUAGGA ...((.((((((((((((((.(((((((((((.(((((....((.(((((.......))))).))....))))).))))).))))))))))))))).)))))((....)).......)). ( -43.60) >DroYak_CAF1 35929 120 - 1 AUUCCAGGAUUGGGAAUUCCAUGGUUGGGAAUGCCAGGAUUCGGGAUUCCAUCAUUAGGAAUGCCACGGUUGGGAAUUCCAUGAUUAGGGAUUCCAGGAUUGGGAAUUCCAUGAUUGGGG ..(((((.....((((((((.((((.......)))).((((((((((((((((((..(((((.(((....)))..))))))))))..)))))))).))))).))))))))....))))). ( -45.30) >DroAna_CAF1 31408 120 - 1 AUGCCACGAUUCGGAAUGCCAUGAUUGGGGAUACCAGGAUUGGGAAUUCCAUGAUUGGGGAUACCAGGAUUGGGAAUACCGUCAAUGGGAAUUCCAGGGUUAGGAAUGCCGUCAUUGGGA ...((.(((((((((((.(((...((((.....))))...)))..))))).....(((.....)))))))))))....(((...((((..(((((.......))))).))))...))).. ( -37.00) >DroPer_CAF1 41833 120 - 1 AUGCCAGGAUUUGGGAUUCCUUCGUUGGGAAUGCCAGGAUUCGGUAUUCCUACAUUGGGGAUGCCAGGAUUUGGGAUUCCUUCGUUGGGAAUGCCAGGAUUCGGUAUUCCUACAUUGGGG (((((.((((((((.((((((.....)))))).))).)))))))))).((((...((((((((((.((((((((.((((((.....)))))).))).)))))))))))))))...)))). ( -53.00) >consensus AUGCCAGGAUUCGGAAUUCCAUGAUUGGGAAUGCCAGGAUUCGGGAUUCCAUCAUUAGGAAUGCCACGAUUUGGAAUUCCAUGAUUAGGGAUUCCAGGAUUGGGAAUUCCAUCAUUGGGA ...((.((((((((.(((((.......))))).)).))))))((((((((.......((....)).((((((((((((((.......))))))))).))))))))))))).......)). (-33.77 = -34.30 + 0.53)
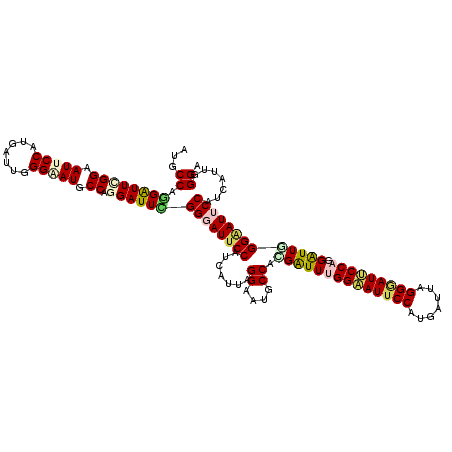
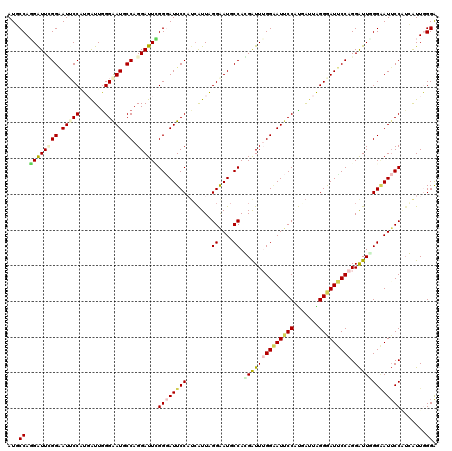
| Location | 14,199,218 – 14,199,314 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 80.82 |
| Mean single sequence MFE | -23.78 |
| Consensus MFE | -14.88 |
| Energy contribution | -15.88 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14199218 96 + 23771897 GGAAUUCCAAACCGUGGCAUUCCUAAUGAUGGAAUCCCGAAUCCUGGCAUUCCCAAUCACGGUAUUCCCAACCCUGGAAUCCCUAAUCAUGGAAUU---- ..(((((((.((((((((((.....)))..(((((.(((.....))).)))))...)))))))(((((.......))))).........)))))))---- ( -25.10) >DroPse_CAF1 33258 96 + 1 GGAAUACCAAAUCCUGGCAUUCCCAACGAAGGAAUCCCAAAUCCUGGCAUCCCCAAUGAAGGAAUACCGAAUCCUGGCAUCCCCAAUGAAGGAAUA---- (((........)))(((.(((((.......))))).)))..((((..(((.....(((.((((........))))..))).....))).))))...---- ( -20.10) >DroSec_CAF1 32966 90 + 1 GGAAUUCCAAACCGUGGCAUUCCUAAUGAUGGAAUCCCUAAUCCUGGCAUUCCUAAUCAUGGAAUUCCAAACCGUGGCAUUCCUAA----------UGAU (((((.(((.....(((.(((((..((((((((((.((.......)).)))))..)))))))))).))).....))).)))))...----------.... ( -29.60) >DroEre_CAF1 34133 96 + 1 GGAAUUCCAAACCGCGGCAUUCCUAAUGAUGGAAUCCCGAAUCCUGGCAUUCCCAAUCAUGGAAUUCCCAAUCCUGGAAUCCCUAAUCAUGGAAUU---- ((.((((((......((.(((((..((((((((((.(((.....))).)))))..)))))))))).))......)))))).)).............---- ( -26.00) >DroYak_CAF1 35969 96 + 1 GGAAUUCCCAACCGUGGCAUUCCUAAUGAUGGAAUCCCGAAUCCUGGCAUUCCCAACCAUGGAAUUCCCAAUCCUGGAAUCCCUAAUCACGGAAUC---- ((((((((....((.((.(((((.......))))))))).....(((.....))).....))))))))...(((..((........))..)))...---- ( -23.30) >DroAna_CAF1 31448 96 + 1 GGUAUUCCCAAUCCUGGUAUCCCCAAUCAUGGAAUUCCCAAUCCUGGUAUCCCCAAUCAUGGCAUUCCGAAUCGUGGCAUUCCUAAUGAUGGAAUC---- ...(((((......(((.....)))((((((((((..(((....(((.....(((....)))....))).....))).)))))..)))))))))).---- ( -18.60) >consensus GGAAUUCCAAACCGUGGCAUUCCUAAUGAUGGAAUCCCGAAUCCUGGCAUUCCCAAUCAUGGAAUUCCCAAUCCUGGAAUCCCUAAUCAUGGAAUC____ (((((.(((.....(((.(((((.......))))).))).....))).))))).......((.((.((.......)).)).))................. (-14.88 = -15.88 + 1.00)
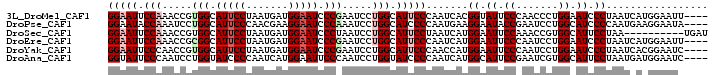
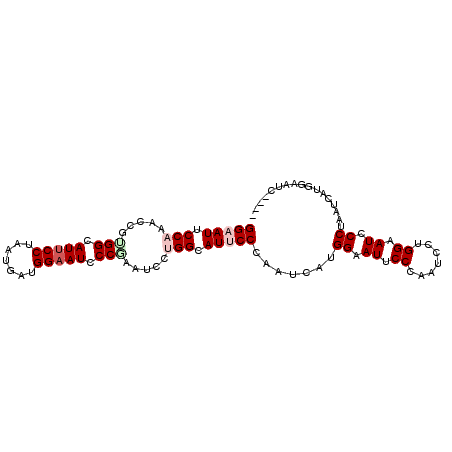
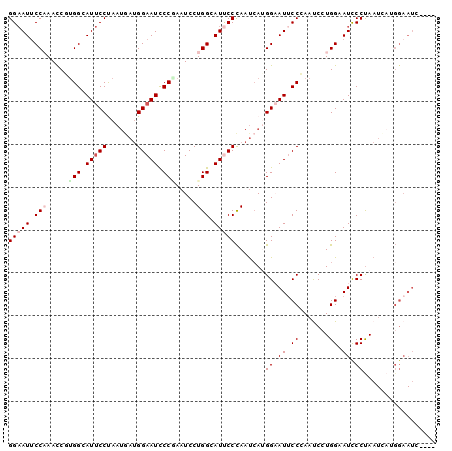
| Location | 14,199,218 – 14,199,314 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 80.82 |
| Mean single sequence MFE | -33.08 |
| Consensus MFE | -27.66 |
| Energy contribution | -27.22 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.962849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14199218 96 - 23771897 ----AAUUCCAUGAUUAGGGAUUCCAGGGUUGGGAAUACCGUGAUUGGGAAUGCCAGGAUUCGGGAUUCCAUCAUUAGGAAUGCCACGGUUUGGAAUUCC ----(((((((......((.(((((..(....(((((.(((...((((.....))))....))).)))))....)..))))).))......))))))).. ( -31.10) >DroPse_CAF1 33258 96 - 1 ----UAUUCCUUCAUUGGGGAUGCCAGGAUUCGGUAUUCCUUCAUUGGGGAUGCCAGGAUUUGGGAUUCCUUCGUUGGGAAUGCCAGGAUUUGGUAUUCC ----.............((((((((((.(((((((((((((.....((((((.((((...)))).)))))).....))))))))).)))))))))))))) ( -40.90) >DroSec_CAF1 32966 90 - 1 AUCA----------UUAGGAAUGCCACGGUUUGGAAUUCCAUGAUUAGGAAUGCCAGGAUUAGGGAUUCCAUCAUUAGGAAUGCCACGGUUUGGAAUUCC ....----------...(((((.(((.(.(.(((.((((((((((..(((((.((.......)).))))))))))..))))).))).).).))).))))) ( -28.90) >DroEre_CAF1 34133 96 - 1 ----AAUUCCAUGAUUAGGGAUUCCAGGAUUGGGAAUUCCAUGAUUGGGAAUGCCAGGAUUCGGGAUUCCAUCAUUAGGAAUGCCGCGGUUUGGAAUUCC ----.(((((.((((((((((((((.......)))))))).))))))))))).(((((...(((.(((((.......))))).)))...)))))...... ( -34.70) >DroYak_CAF1 35969 96 - 1 ----GAUUCCGUGAUUAGGGAUUCCAGGAUUGGGAAUUCCAUGGUUGGGAAUGCCAGGAUUCGGGAUUCCAUCAUUAGGAAUGCCACGGUUGGGAAUUCC ----.............((((((((..(((((((.((((((((((..(((((.((.......)).))))))))))..))))).)).))))).)))))))) ( -33.50) >DroAna_CAF1 31448 96 - 1 ----GAUUCCAUCAUUAGGAAUGCCACGAUUCGGAAUGCCAUGAUUGGGGAUACCAGGAUUGGGAAUUCCAUGAUUGGGGAUACCAGGAUUGGGAAUACC ----.(((((.......((....)).(((((((((((.(((...((((.....))))...)))..))))).....(((.....))))))))))))))... ( -29.40) >consensus ____AAUUCCAUGAUUAGGGAUGCCAGGAUUGGGAAUUCCAUGAUUGGGAAUGCCAGGAUUCGGGAUUCCAUCAUUAGGAAUGCCACGGUUUGGAAUUCC .................((((((((.((((((((.(((((.((((..(((((.((.......)).)))))...))))))))).)).)))))))))))))) (-27.66 = -27.22 + -0.44)

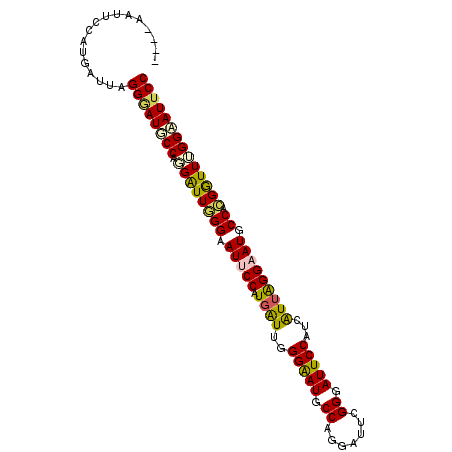

| Location | 14,199,258 – 14,199,348 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 81.85 |
| Mean single sequence MFE | -21.27 |
| Consensus MFE | -13.79 |
| Energy contribution | -14.46 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.839859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14199258 90 + 23771897 AUCCUGGCAUUCCCAAUCACGGUAUUCCCAACCCUGGAAUCCCUAAUCAUGGAAUUCCAAACCGCGGCAUUCCUAAUGAUGGAAUCCCUA .....((.(((((.......((.(((((.......))))).))..((((((((((.((.......)).)))))..)))))))))).)).. ( -23.00) >DroPse_CAF1 33298 90 + 1 AUCCUGGCAUCCCCAAUGAAGGAAUACCGAAUCCUGGCAUCCCCAAUGAAGGAAUACCGAAUCCUGGCAUCCCCAAUGUAGGAAUACCGA .(((((.(((.....(((.((((........((((..(((.....))).))))........))))..))).....))))))))....... ( -17.19) >DroEre_CAF1 34173 90 + 1 AUCCUGGCAUUCCCAAUCAUGGAAUUCCCAAUCCUGGAAUCCCUAAUCAUGGAAUUCCAAACCGCGGCAUUCCUAAUGAUGGAAUCCCGA ....(((.(((((..((((((((((.((......((((((.((.......)).))))))......)).)))))..)))))))))).))). ( -26.40) >DroYak_CAF1 36009 90 + 1 AUCCUGGCAUUCCCAACCAUGGAAUUCCCAAUCCUGGAAUCCCUAAUCACGGAAUCCCCAACCGCGGCAUUCCUAAUGAUGGAAUCCCUA .....((.(((((.......((.(((((.......))))).))..((((.(((((.(((....).)).)))))...))))))))).)).. ( -21.30) >DroAna_CAF1 31488 90 + 1 AUCCUGGUAUCCCCAAUCAUGGCAUUCCGAAUCGUGGCAUUCCUAAUGAUGGAAUCCCCAAUCCUGGUAUUCCUAACCAUGGCAUUCCCA ..((((((.......(((((((.((.(((.....))).)).))..)))))(((((..(((....))).)))))..)))).))........ ( -17.30) >DroPer_CAF1 41913 90 + 1 AUCCUGGCAUUCCCAACGAAGGAAUCCCAAAUCCUGGCAUCCCUAAUGUAGGAAUACCGAAUCCUGGCAUUCCCAACGAAGGAAUCCCAA ....(((.(((((...((..(((((.(((.....(((.((.((((...)))).)).))).....))).)))))...))..))))).))). ( -22.40) >consensus AUCCUGGCAUUCCCAAUCAUGGAAUUCCCAAUCCUGGAAUCCCUAAUCAUGGAAUACCAAACCCCGGCAUUCCUAAUGAUGGAAUCCCGA .....((.(((((.......((.((.((.......)).)).)).......))))).)).......((.(((((.......))))).)).. (-13.79 = -14.46 + 0.67)

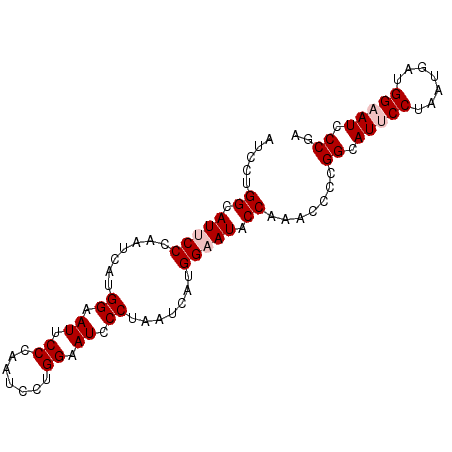
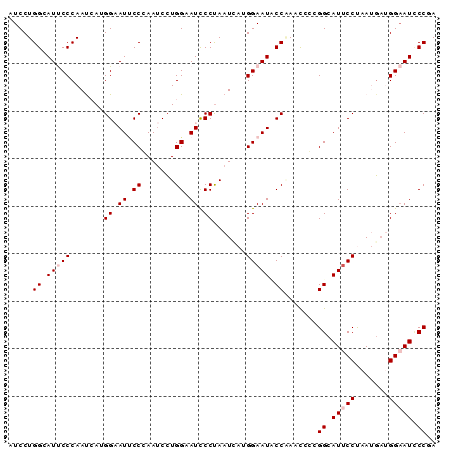
| Location | 14,199,258 – 14,199,348 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 81.85 |
| Mean single sequence MFE | -37.05 |
| Consensus MFE | -32.40 |
| Energy contribution | -30.90 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.44 |
| Mean z-score | -4.55 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14199258 90 - 23771897 UAGGGAUUCCAUCAUUAGGAAUGCCGCGGUUUGGAAUUCCAUGAUUAGGGAUUCCAGGGUUGGGAAUACCGUGAUUGGGAAUGCCAGGAU ..((.(((((.......))))).))....(((((.(((((.((((((.(((((((.......))))).)).))))))))))).))))).. ( -31.90) >DroPse_CAF1 33298 90 - 1 UCGGUAUUCCUACAUUGGGGAUGCCAGGAUUCGGUAUUCCUUCAUUGGGGAUGCCAGGAUUCGGUAUUCCUUCAUUGGGGAUGCCAGGAU ..((((((((((....(((((((((.(((((((((((((((.....))))))))).)))))))))))))))....))))))))))..... ( -47.40) >DroEre_CAF1 34173 90 - 1 UCGGGAUUCCAUCAUUAGGAAUGCCGCGGUUUGGAAUUCCAUGAUUAGGGAUUCCAGGAUUGGGAAUUCCAUGAUUGGGAAUGCCAGGAU ..((.((((((((((..(((((.((.((((((((((((((.......))))))))).))))))).))))))))))..))))).))..... ( -36.40) >DroYak_CAF1 36009 90 - 1 UAGGGAUUCCAUCAUUAGGAAUGCCGCGGUUGGGGAUUCCGUGAUUAGGGAUUCCAGGAUUGGGAAUUCCAUGGUUGGGAAUGCCAGGAU ..((.((((((((((..(((((.((.(((((.((((((((.......))))))))..))))))).))))))))))..))))).))..... ( -35.40) >DroAna_CAF1 31488 90 - 1 UGGGAAUGCCAUGGUUAGGAAUACCAGGAUUGGGGAUUCCAUCAUUAGGAAUGCCACGAUUCGGAAUGCCAUGAUUGGGGAUACCAGGAU ..((....((.((((.......))))((((((((.(((((.......))))).)).))))))))....))....((((.....))))... ( -27.70) >DroPer_CAF1 41913 90 - 1 UUGGGAUUCCUUCGUUGGGAAUGCCAGGAUUCGGUAUUCCUACAUUAGGGAUGCCAGGAUUUGGGAUUCCUUCGUUGGGAAUGCCAGGAU ((((.((((((.((..((((((.((((.((((((((((((((...)))))))))).)))))))).)))))).))..)))))).))))... ( -43.50) >consensus UAGGGAUUCCAUCAUUAGGAAUGCCACGAUUCGGAAUUCCAUCAUUAGGGAUGCCAGGAUUGGGAAUUCCAUGAUUGGGAAUGCCAGGAU ..((.(((((...(((((((((.((.((((((((((((((.......)))))))).)))))))).))))).))))..))))).))..... (-32.40 = -30.90 + -1.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:32 2006