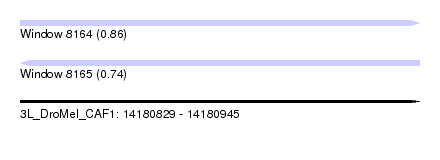
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,180,829 – 14,180,945 |
| Length | 116 |
| Max. P | 0.862453 |

| Location | 14,180,829 – 14,180,945 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 77.43 |
| Mean single sequence MFE | -32.09 |
| Consensus MFE | -15.90 |
| Energy contribution | -16.02 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
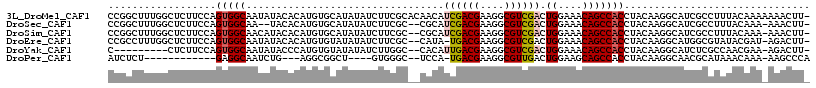
>3L_DroMel_CAF1 14180829 116 + 23771897 CCGGCUUUGGCUCUUCCAGUGGCAAUAUACACAUGUGCAUAUAUCUUCGCACAACAUCGACGAAGGCGUCGACUGGAAACAGCCACCUACAAGGCAUCGCCUUUACAAAAAAACUU- ..(((..(((((.(((((((.............(((((..........)))))....(((((....))))))))))))..)))))((.....))....)))...............- ( -33.50) >DroSec_CAF1 15102 111 + 1 CCGGCUUUGGCUCUUCCAGUGGCAA--UACACAUGUGCAUAUAUCUUCGC--CGCAUCGACGAAGGCGUCGACUGGAAACAGCCACCUACAAGGCAUCGCCUUUACAAA-AAACUU- ..(((..(((((.((((((((((..--.....................))--))).((((((....)))))).)))))..)))))((.....))....)))........-......- ( -31.70) >DroSim_CAF1 15143 113 + 1 CCGGCUUUGGCUCUUCCAGUGGCAACAUACACAUGUGCAUAUAUCUUCGC--CGCAUCGACGAAGGCGUCGACUGGAAACAGCCACCUACAAGGCAUCGCCUUUACAAA-AAACUU- ..(((..(((((.((((((((((.((((....))))............))--))).((((((....)))))).)))))..)))))((.....))....)))........-......- ( -32.32) >DroEre_CAF1 15920 112 + 1 CCGCCUUUGGCUCUUCCAGUGGCAAUAUACACAUGUGUAUAUAUCUUCGC--CAUA-UGACGAAGGCGUCGACUGGAAACAGCCACCUACAAGGCAUGGCGUAUACGAU-AGACUU- .((((..(((((.((((((((((.(((((((....))))))).((((((.--(...-.).)))))).))).)))))))..)))))((.....))...))))........-......- ( -37.90) >DroYak_CAF1 17619 104 + 1 C---------CUCUUCCAGUGGCAAUAUACCCAUGUGUAUAUAUCUUGGC--CACAUUGACGAAGGCGUCGACUGGAAACAGCCACCUACAAGGCAUCUCGCCAACGAA-AGACUU- .---------.((((.(..((((.((((((......))))))......((--(...((((((....))))))(((....)))..........))).....))))..).)-)))...- ( -28.70) >DroPer_CAF1 23868 94 + 1 AUCUCU------------GAGGCAAUCUG---AGGCGGCU----GUGGGC--UCCA-UGACGAAGGCGUUGACUGGAAGCAGCCACCUACAAGGCAACGCAUAAACAAA-AAGCCCA ......------------(.(((....((---(((.((((----((....--((((-(((((....)))))..)))).)))))).))).)).(....)...........-..)))). ( -28.40) >consensus CCGGCUUUGGCUCUUCCAGUGGCAAUAUACACAUGUGCAUAUAUCUUCGC__CACAUCGACGAAGGCGUCGACUGGAAACAGCCACCUACAAGGCAUCGCCUUUACAAA_AAACUU_ ..................(((((.................................((((((....)))))).((....)))))))............................... (-15.90 = -16.02 + 0.11)
| Location | 14,180,829 – 14,180,945 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.43 |
| Mean single sequence MFE | -38.45 |
| Consensus MFE | -16.93 |
| Energy contribution | -19.95 |
| Covariance contribution | 3.02 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.735035 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
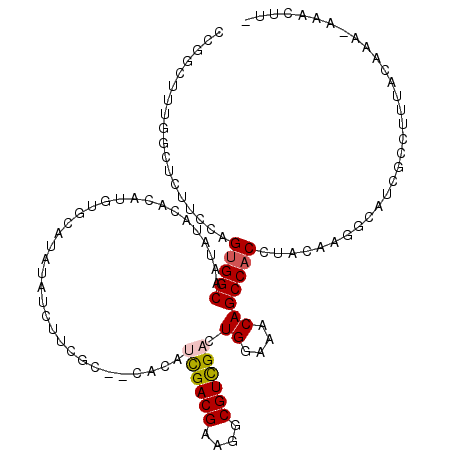
>3L_DroMel_CAF1 14180829 116 - 23771897 -AAGUUUUUUUGUAAAGGCGAUGCCUUGUAGGUGGCUGUUUCCAGUCGACGCCUUCGUCGAUGUUGUGCGAAGAUAUAUGCACAUGUGUAUAUUGCCACUGGAAGAGCCAAAGCCGG -...............(((...(((.....)))((((.(((((((((((((....)))))))...(((....(((((((((....)))))))))..)))))))))))))...))).. ( -38.70) >DroSec_CAF1 15102 111 - 1 -AAGUUU-UUUGUAAAGGCGAUGCCUUGUAGGUGGCUGUUUCCAGUCGACGCCUUCGUCGAUGCG--GCGAAGAUAUAUGCACAUGUGUA--UUGCCACUGGAAGAGCCAAAGCCGG -......-........(((...(((.....)))((((.(((((((((((((....))))))...(--((((..((((((....)))))).--))))).)))))))))))...))).. ( -40.90) >DroSim_CAF1 15143 113 - 1 -AAGUUU-UUUGUAAAGGCGAUGCCUUGUAGGUGGCUGUUUCCAGUCGACGCCUUCGUCGAUGCG--GCGAAGAUAUAUGCACAUGUGUAUGUUGCCACUGGAAGAGCCAAAGCCGG -......-........(((...(((.....)))((((.(((((((((((((....))))))...(--((((..((((((....))))))...))))).)))))))))))...))).. ( -42.50) >DroEre_CAF1 15920 112 - 1 -AAGUCU-AUCGUAUACGCCAUGCCUUGUAGGUGGCUGUUUCCAGUCGACGCCUUCGUCA-UAUG--GCGAAGAUAUAUACACAUGUGUAUAUUGCCACUGGAAGAGCCAAAGGCGG -......-........((((..(((.....)))((((.((((((((.((((....)))).-...(--((...(((((((((....))))))))))))))))))))))))...)))). ( -42.90) >DroYak_CAF1 17619 104 - 1 -AAGUCU-UUCGUUGGCGAGAUGCCUUGUAGGUGGCUGUUUCCAGUCGACGCCUUCGUCAAUGUG--GCCAAGAUAUAUACACAUGGGUAUAUUGCCACUGGAAGAG---------G -...(((-((((.((((((.((((((.......(((((....)))))((((....)))).(((((--.............))))))))))).)))))).))))))).---------. ( -35.82) >DroPer_CAF1 23868 94 - 1 UGGGCUU-UUUGUUUAUGCGUUGCCUUGUAGGUGGCUGCUUCCAGUCAACGCCUUCGUCA-UGGA--GCCCAC----AGCCGCCU---CAGAUUGCCUC------------AGAGAU .((((..-((((....((((......))))((((((((.(((((....(((....)))..-))))--)....)----))))))).---))))..)))).------------...... ( -29.90) >consensus _AAGUUU_UUUGUAAAGGCGAUGCCUUGUAGGUGGCUGUUUCCAGUCGACGCCUUCGUCAAUGUG__GCGAAGAUAUAUGCACAUGUGUAUAUUGCCACUGGAAGAGCCAAAGCCGG ......................(((.....)))((((.((((((((.((((....))))........((((..((((((....))))))...)))).))))))))))))........ (-16.93 = -19.95 + 3.02)

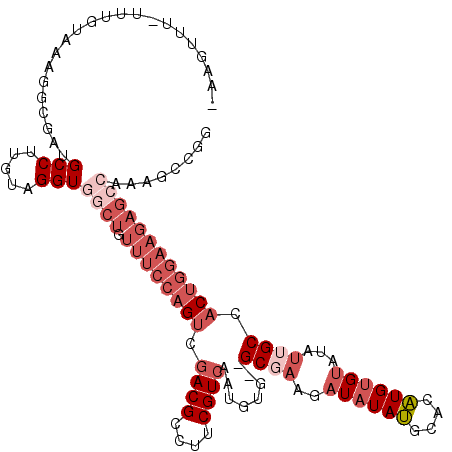
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:21 2006