| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,174,309 – 14,174,418 |
| Length | 109 |
| Max. P | 0.886692 |

| Location | 14,174,309 – 14,174,418 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 63.21 |
| Mean single sequence MFE | -21.27 |
| Consensus MFE | -9.45 |
| Energy contribution | -10.15 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14174309 109 - 23771897 CCC-----CCCUCUCCUCAUCG-UAAGUCACCUGACAAAGGGACUCGUCUCAUAACGAGUUAACUCUUUACACAGAAGUGG---CAACCAGUUUAACGUAUGUACAUACAUAUGUAUA ...-----..............-...((((((((..((((((((((((......))))))...))))))...)))..))))---)..........((((((((....))))))))... ( -25.60) >DroSec_CAF1 8734 99 - 1 CC------CCCUCUCCUUAUCU-UAAGUCACCUGACAAAGGGACUCGUCUCAUAACGAGUUAACUCUUUACACAGAAGUGG---CAACCAGUUUGACGUAC---------UACGUACA ..------..............-...((((.(((..((((((((((((......))))))...)))))).(((....))).---....)))..))))((((---------...)))). ( -21.00) >DroSim_CAF1 8808 100 - 1 CCG-----CCCUCUCCUCAUCU-UAAGUCACCUGACAAAGGGACUCGUCUCAUAACGAGUUAACUCUUUACACAGAAGUGG---CAACCAGUUUGACGUAC---------UACGUACA ..(-----((((((........-...(((....)))((((((((((((......))))))...))))))....)).)).))---)............((((---------...)))). ( -21.40) >DroWil_CAF1 13105 96 - 1 AUA-----------CAUUAACUUAAAGUCACCUGACAAAGGGUCGCCUCUUAUUGGGUCUUAAGU--------AGAAGCUUUUGUUGC--CUUUAAC-UAUCUCAAUGCAGGUGGAUA ...-----------.............(((((((.(.(((((....)))))((((((...((..(--------(((.((.......))--.))))..-)).))))))))))))))... ( -20.60) >DroYak_CAF1 11065 94 - 1 CCCCUUCUCCCUCCCCUCAUCC-CAAGGCACCUGACAGAGGAACUCGUCUCAUAACGAGUUAACUCUUUACGCAGAAGUGG---CAACCAGUUUGCCG-------------------- ...(((((.(....((((....-..((....))....))))(((((((......)))))))..........).))))).((---(((.....))))).-------------------- ( -22.90) >DroAna_CAF1 7847 93 - 1 UCUCA-CAAGCACUCUUAAAGU-AAAGUCACCUGACAAUGGGAACCUCCUCAUAACAAGUUGAAUCUUUACACAGAAAUAU---CAACCUGUUUGACU-------------------- .....-((((((........((-(((((((.((....(((((......)))))....)).)))..))))))...((....)---)....))))))...-------------------- ( -16.10) >consensus CCC_____CCCUCUCCUCAUCU_UAAGUCACCUGACAAAGGGACUCGUCUCAUAACGAGUUAACUCUUUACACAGAAGUGG___CAACCAGUUUGACGUA__________UA_GUA_A ..........................(((((((.....)))(((((((......)))))))................................))))..................... ( -9.45 = -10.15 + 0.70)


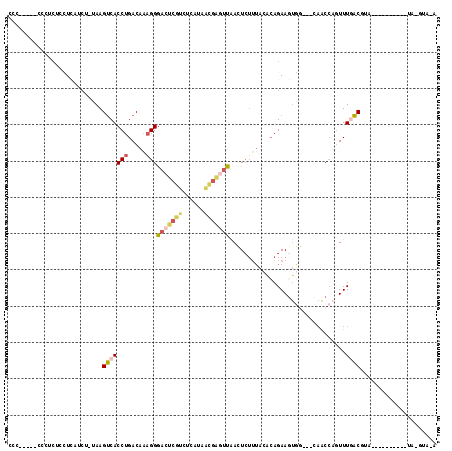
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:19 2006