
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,161,236 – 14,161,384 |
| Length | 148 |
| Max. P | 0.999476 |

| Location | 14,161,236 – 14,161,345 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 76.44 |
| Mean single sequence MFE | -22.33 |
| Consensus MFE | -16.49 |
| Energy contribution | -16.17 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.74 |
| SVM decision value | 3.64 |
| SVM RNA-class probability | 0.999476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
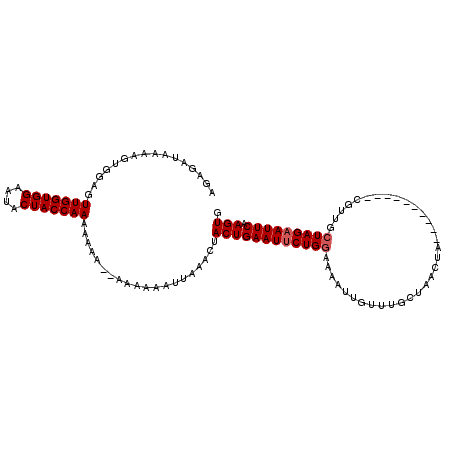
>3L_DroMel_CAF1 14161236 109 + 23771897 GCUAAUUUAGUAGAUUAGCUUAUUUGAGGGUUAAGUGAGAGAUAAAAGUAGAGUUGGUGGAAUACUACCAAAAAAA--AAAAAAUUAAACUACUGAAUUCUGGAAAAUUGC ...((((((((((.(((((((((((((...)))))))))..............(((((((....))))))).....--......)))).))))))))))............ ( -22.40) >DroSim_CAF1 162056 107 + 1 GCUAAUUUAGUAGAUUAGCUCAUUUGAGAGUUAAGUGAGAGAUAAAAGUGGAGUUGGUGGAAUACUACCAAAAAU----AAUAAUUAAACGACUGAAUUCUGAAAAAUUGU (((((((.....)))))))...(((.((((((.(((............((...(((((((....)))))))...)----)...........))).)))))).)))...... ( -21.80) >DroEre_CAF1 159853 89 + 1 GCUA-UUUAGUAGAUUAGCUUAUUUGGCUGUUAAGUGAGAGAUAAAAGUGGAGUUGGUGGAAUACCACCAAAAAUUCACAAUAUUUAUAC--------------------- ((((-.((....)).))))...(((.(((....))).))).......(((((.(((((((....)))))))...)))))...........--------------------- ( -22.80) >consensus GCUAAUUUAGUAGAUUAGCUUAUUUGAGAGUUAAGUGAGAGAUAAAAGUGGAGUUGGUGGAAUACUACCAAAAAU____AAUAAUUAAAC_ACUGAAUUCUG_AAAAUUG_ (((((((.....)))))))((((((((...))))))))...............(((((((....)))))))........................................ (-16.49 = -16.17 + -0.33)



| Location | 14,161,273 – 14,161,384 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 84.10 |
| Mean single sequence MFE | -18.49 |
| Consensus MFE | -14.18 |
| Energy contribution | -14.84 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14161273 111 + 23771897 AGAGAUAAAAGUAGAGUUGGUGGAAUACUACCAAAAAAA--AAAAAAUUAAACUACUGAAUUCUGGAAAAUUGCCUGCUAAAUAUUCAUUCAAGCAUUGCUAGCAUUCAAGUG .........(((((..(((((((....))))))).....--...........)))))((((.((((.....(((.((.............)).)))...)))).))))..... ( -17.79) >DroSec_CAF1 158934 103 + 1 AGAGAUAAAAGUGGAGUUGGUGGAAUACUACCAAAAAAAAAAAAAAAUAAAACUACUGAAUUCUGGAAAAUUGUUUGCAAACUA----------CGUUGCUAGAAUUCAAGUG .........(((((..(((((((....)))))))..................)))))(((((((((......(((....)))..----------.....)))))))))..... ( -20.07) >DroSim_CAF1 162093 99 + 1 AGAGAUAAAAGUGGAGUUGGUGGAAUACUACCAAAAAU----AAUAAUUAAACGACUGAAUUCUGAAAAAUUGUUUGCUAACUA----------CGUUGCUAGAAUUCAAGUG ................(((((((....)))))))....----............(((((((((((..((...(((....)))..----------..))..)))))))).))). ( -17.60) >consensus AGAGAUAAAAGUGGAGUUGGUGGAAUACUACCAAAAAAA__AAAAAAUUAAACUACUGAAUUCUGGAAAAUUGUUUGCUAACUA__________CGUUGCUAGAAUUCAAGUG ................(((((((....)))))))....................((((((((((((.................................))))))))).))). (-14.18 = -14.84 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:14 2006