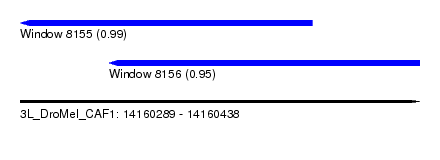
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,160,289 – 14,160,438 |
| Length | 149 |
| Max. P | 0.985554 |

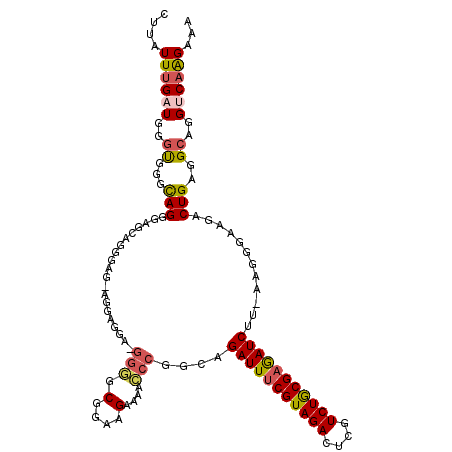
| Location | 14,160,289 – 14,160,398 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 87.91 |
| Mean single sequence MFE | -36.73 |
| Consensus MFE | -28.12 |
| Energy contribution | -28.40 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14160289 109 - 23771897 GGGGCGGAAGAAAACCCGGCAGAUUUCGUAGACUCGUCUGCGAGAUCUU-AAGGAAAGACUGAGGCAGGUCAAGAAAAACAGGCCGGAAAACACAGUUC-------UGGCUAGAGAG (((.(....)....)))...((((((((((((....)))))))))))).-.......((((......))))..........((((((((.......)))-------)))))...... ( -37.40) >DroSec_CAF1 157925 115 - 1 GGGGCGGAAGAAAACCCGGCAGAUUUCGUAGACUCGUCUGCGAGAUCUU-AAG-GAAGACUGAGGCAGGUCAAGAAAAUCAGGUCGAAAAACACAGUUCUGGGUUCUGGCUAGAGAG .((.((((.....((((((.((((((((((((....)))))))))))).-...-...(((((.(...(..(..........)..)......).))))))))))))))).))...... ( -34.60) >DroSim_CAF1 161087 109 - 1 GGGGCGGAAGAAAACCCGGCAGAUUUCGUAGACGCGUCUACGAGAUCUU-AAGGGAAGACUGAGGCAGGUCAAGAAAAUCAGGCCGGAAAACACAGUUC-------UGGCUGGAGAG ....(....)....(((...((((((((((((....)))))))))))).-..)))..((((......)))).......((.((((((((.......)))-------))))).))... ( -39.60) >DroEre_CAF1 158855 107 - 1 ---GCGGAAGAAAAGCCGGCAGAUUUCGUAGACUUGUCUGCGAAAUCUGAAAGGGAAGACUGAGGCAGGUCAGGAAAAUCAGCCCGGAAAACACAGUUC-------UGGCUAGAGGG ---.(....)...(((((((((((((((((((....)))))))))))))...(((..((((((......)))).....))..))).............)-------)))))...... ( -36.50) >DroYak_CAF1 165719 110 - 1 GGUGCGGAAGAAAAGCCGGCAGAUUCCGUAGACUUGUCUGCGAGAUCGCGAAGGGAAGACUGAGGCAGGUCAGGAAAAUCAGGCCGGAAAACACAGUUU-------UGGCUCGAGGG .(.((.((((.....(((((.((((((...((((((((((((....)))..((......)).))))))))).)))..)))..))))).........)))-------).)).)..... ( -35.54) >consensus GGGGCGGAAGAAAACCCGGCAGAUUUCGUAGACUCGUCUGCGAGAUCUU_AAGGGAAGACUGAGGCAGGUCAAGAAAAUCAGGCCGGAAAACACAGUUC_______UGGCUAGAGAG (((.(....)....)))(((.(((((((((((....)))))))))))..........(((((.(...((((..........))))......).)))))..........)))...... (-28.12 = -28.40 + 0.28)

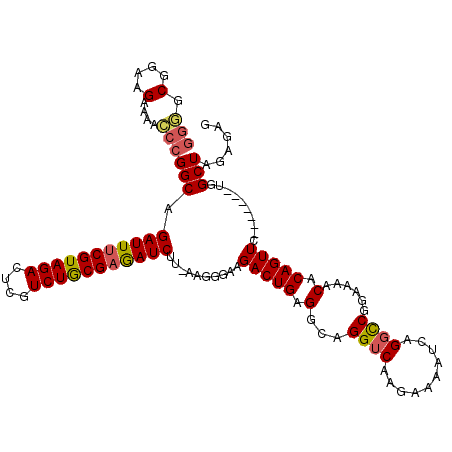
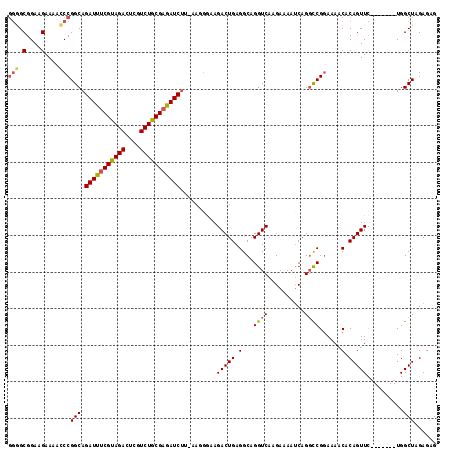
| Location | 14,160,322 – 14,160,438 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.96 |
| Mean single sequence MFE | -34.54 |
| Consensus MFE | -24.68 |
| Energy contribution | -25.08 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14160322 116 - 23771897 CUUAUUUGAUGGGUGUGUAGGGAGCAGGGAGGAGGAGGAGGGGGCGGAAGAAAACCCGGCAGAUUUCGUAGACUCGUCUGCGAGAUCUU-AAGGAAAGACUGAGGCAGGUCAAGAAA ....((((((..((.(.(((...................(((..(....)....)))...((((((((((((....)))))))))))).-.........)))).))..))))))... ( -31.70) >DroSec_CAF1 157965 112 - 1 CUUAUUUGAUGGGUGGGUAGGGAGCAGGGAGCAGGAG---GGGGCGGAAGAAAACCCGGCAGAUUUCGUAGACUCGUCUGCGAGAUCUU-AAG-GAAGACUGAGGCAGGUCAAGAAA ....((((((..((...(((..........((....(---((..(....)....))).))((((((((((((....)))))))))))).-...-.....)))..))..))))))... ( -33.50) >DroSim_CAF1 161120 114 - 1 CUUAUUUGAUGGGUG--CAGUGAGCAGGGAGGAGGAGGAAGGGGCGGAAGAAAACCCGGCAGAUUUCGUAGACGCGUCUACGAGAUCUU-AAGGGAAGACUGAGGCAGGUCAAGAAA ....((((((..((.--((((.......................(....)....(((...((((((((((((....)))))))))))).-..)))...))))..))..))))))... ( -34.60) >DroEre_CAF1 158888 109 - 1 CUUGUUUGUUGGGCGGGCAGGGCGAAGGC----UGGGGG----GCGGAAGAAAAGCCGGCAGAUUUCGUAGACUUGUCUGCGAAAUCUGAAAGGGAAGACUGAGGCAGGUCAGGAAA ((((((((((..((.(((..(((....))----).....----.(....)....))).))((((((((((((....))))))))))))...............)))))).))))... ( -39.30) >DroYak_CAF1 165752 113 - 1 CUUAUUCGUUGGAUGGGCAGGGCGGAGAA----UGAGGGGGGUGCGGAAGAAAAGCCGGCAGAUUCCGUAGACUUGUCUGCGAGAUCGCGAAGGGAAGACUGAGGCAGGUCAGGAAA ((((((.....))))))....((((((..----((..((.....(....).....))..))..)))))).((((((((((((....)))..((......)).)))))))))...... ( -33.60) >consensus CUUAUUUGAUGGGUGGGCAGGGAGCAGGGAG_AGGAGGA_GGGGCGGAAGAAAACCCGGCAGAUUUCGUAGACUCGUCUGCGAGAUCUU_AAGGGAAGACUGAGGCAGGUCAAGAAA ....((((((..((...(((....................(((.(....)....)))....(((((((((((....)))))))))))............)))..))..))))))... (-24.68 = -25.08 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:12 2006