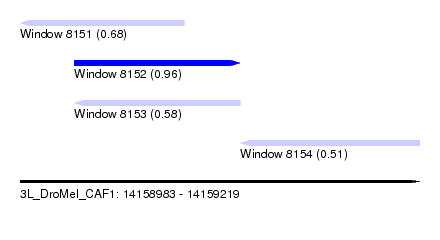
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,158,983 – 14,159,219 |
| Length | 236 |
| Max. P | 0.962328 |

| Location | 14,158,983 – 14,159,080 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 91.94 |
| Mean single sequence MFE | -21.50 |
| Consensus MFE | -17.28 |
| Energy contribution | -17.36 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14158983 97 - 23771897 GCAUUUGAAUGCAUUUCAAUUUGAGUCGCUCAUUCAGUGAGUCGUUCGAUCAACGAGUUUUUUUUU-------CAGGUUGUUGUUUAAACCAAUUAAAUGUCAA ((((((((.((...........((.(((((.....))))).))(((..(.((((((.((.......-------.)).)))))).)..))))).))))))))... ( -20.40) >DroSec_CAF1 156671 94 - 1 GCAUUUGAAUGCAUUUCCAUUUGAGUCGCUCAUUCAGUGAGUCGUUCGAUCAACGAGUUU---UUU-------CAGGUUGUUGUUUAAACCAAUUAAAUGUCAA ((((((((.((...........((.(((((.....))))).))(((..(.((((((.((.---...-------.)).)))))).)..))))).))))))))... ( -20.40) >DroSim_CAF1 159766 94 - 1 GCAUUUGAAUGCAUUUCCAUUUGAGUCGCUCAUUCAGUGAGUCGUUCGAUCAACGAGUUU---UUU-------CAGGUUGUUGUUUAAACCAAUUAAAUGUCAA ((((((((.((...........((.(((((.....))))).))(((..(.((((((.((.---...-------.)).)))))).)..))))).))))))))... ( -20.40) >DroEre_CAF1 157529 94 - 1 GCAUUUGAAUGCAUUUCCAUUUGAGUCGCUCACUCAGUGGGUCGUUCGAUCAACGAGUUU---UUU-------CAGCUCGUUGUUUAAACCAAUUAAAUGUCAA ((((((((.((....((((((.((((.....))))))))))..(((..(.(((((((((.---...-------.))))))))).)..))))).))))))))... ( -28.00) >DroYak_CAF1 164209 98 - 1 GCAUUUGAAUGCAUUUCCAUUUGAGUCGCUCAUUCAGUGGGUCGUUCGAUCAACGAGUUU---UUUUUUUGUUCAAC---UUGUUUAAACCAAUUAAAUGUCAA ((((((((.((.........(((((..((((((...))))))..)))))..((((((((.---...........)))---))))).....)).))))))))... ( -18.30) >consensus GCAUUUGAAUGCAUUUCCAUUUGAGUCGCUCAUUCAGUGAGUCGUUCGAUCAACGAGUUU___UUU_______CAGGUUGUUGUUUAAACCAAUUAAAUGUCAA ((((((((.((...........((.(((((.....))))).))(((..(.((((((.((...............)).)))))).)..))))).))))))))... (-17.28 = -17.36 + 0.08)

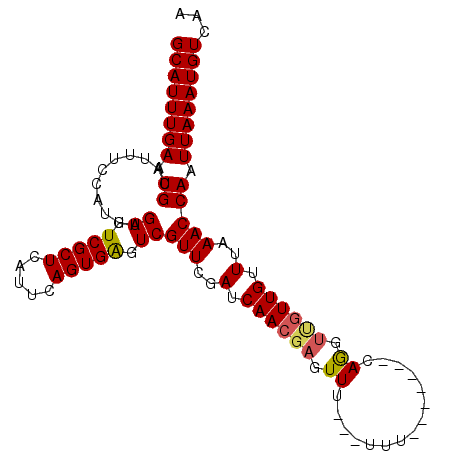

| Location | 14,159,015 – 14,159,113 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 95.69 |
| Mean single sequence MFE | -21.38 |
| Consensus MFE | -19.14 |
| Energy contribution | -19.10 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14159015 98 + 23771897 AAAAAAAACUCGUUGAUCGAACGACUCACUGAAUGAGCGACUCAAAUUGAAAUGCAUUCAAAUGCCAUUCCAUCAAACUGGGUCAAAUGCCGCUU---CUU .........(((((.....)))))((((.....)))).((((((..(((((((((((....))).))))...))))..))))))...........---... ( -17.60) >DroSec_CAF1 156703 95 + 1 AA---AAACUCGUUGAUCGAACGACUCACUGAAUGAGCGACUCAAAUGGAAAUGCAUUCAAAUGCCAUUCCAUCAAACUGGGUCAAAUGCCGCUU---CUU ..---....(((((.....)))))((((.....)))).((((((.((((((..((((....))))..)))))).....))))))...........---... ( -20.40) >DroSim_CAF1 159798 95 + 1 AA---AAACUCGUUGAUCGAACGACUCACUGAAUGAGCGACUCAAAUGGAAAUGCAUUCAAAUGCCAUUCCAUCAAACUGGGUCAAAUGCCGCUU---CUU ..---....(((((.....)))))((((.....)))).((((((.((((((..((((....))))..)))))).....))))))...........---... ( -20.40) >DroEre_CAF1 157561 95 + 1 AA---AAACUCGUUGAUCGAACGACCCACUGAGUGAGCGACUCAAAUGGAAAUGCAUUCAAAUGCCAUUCCAUCAAACUGGGUCAAAUGCCGCUU---CUU ..---....(((.....)))..((((((.(((((.....))))).((((((..((((....))))..)))))).....))))))...........---... ( -24.50) >DroYak_CAF1 164245 98 + 1 AA---AAACUCGUUGAUCGAACGACCCACUGAAUGAGCGACUCAAAUGGAAAUGCAUUCAAAUGCCAUUCCAUCAAACUGGGUCGAAUGCCGCUUCUUCUU ..---....(((.....))).(((((((.(((.((((...))))..(((((..((((....))))..))))))))...)))))))................ ( -24.00) >consensus AA___AAACUCGUUGAUCGAACGACUCACUGAAUGAGCGACUCAAAUGGAAAUGCAUUCAAAUGCCAUUCCAUCAAACUGGGUCAAAUGCCGCUU___CUU .........(((.....)))..((((((.(((.((((...))))..(((((..((((....))))..))))))))...))))))................. (-19.14 = -19.10 + -0.04)


| Location | 14,159,015 – 14,159,113 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 95.69 |
| Mean single sequence MFE | -26.46 |
| Consensus MFE | -22.24 |
| Energy contribution | -22.20 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14159015 98 - 23771897 AAG---AAGCGGCAUUUGACCCAGUUUGAUGGAAUGGCAUUUGAAUGCAUUUCAAUUUGAGUCGCUCAUUCAGUGAGUCGUUCGAUCAACGAGUUUUUUUU (((---((((((........)).(((.(((.(((((((..(((((((...(((.....))).....)))))))...))))))).))))))..))))))).. ( -24.80) >DroSec_CAF1 156703 95 - 1 AAG---AAGCGGCAUUUGACCCAGUUUGAUGGAAUGGCAUUUGAAUGCAUUUCCAUUUGAGUCGCUCAUUCAGUGAGUCGUUCGAUCAACGAGUUU---UU .((---((((((........)).(((.(((.(((((((.((..((((......))))..))(((((.....)))))))))))).))))))..))))---)) ( -25.30) >DroSim_CAF1 159798 95 - 1 AAG---AAGCGGCAUUUGACCCAGUUUGAUGGAAUGGCAUUUGAAUGCAUUUCCAUUUGAGUCGCUCAUUCAGUGAGUCGUUCGAUCAACGAGUUU---UU .((---((((((........)).(((.(((.(((((((.((..((((......))))..))(((((.....)))))))))))).))))))..))))---)) ( -25.30) >DroEre_CAF1 157561 95 - 1 AAG---AAGCGGCAUUUGACCCAGUUUGAUGGAAUGGCAUUUGAAUGCAUUUCCAUUUGAGUCGCUCACUCAGUGGGUCGUUCGAUCAACGAGUUU---UU .((---((((.......((((((....(((((((..((((....))))..)))))))(((((.....))))).))))))..(((.....)))))))---)) ( -28.90) >DroYak_CAF1 164245 98 - 1 AAGAAGAAGCGGCAUUCGACCCAGUUUGAUGGAAUGGCAUUUGAAUGCAUUUCCAUUUGAGUCGCUCAUUCAGUGGGUCGUUCGAUCAACGAGUUU---UU ....((((((......(((((((....(((((((..((((....))))..)))))))(((((.....))))).))))))).(((.....)))))))---)) ( -28.00) >consensus AAG___AAGCGGCAUUUGACCCAGUUUGAUGGAAUGGCAUUUGAAUGCAUUUCCAUUUGAGUCGCUCAUUCAGUGAGUCGUUCGAUCAACGAGUUU___UU ......((((((........)).(((.(((.(((((((.((..((((......))))..))(((((.....)))))))))))).))))))..))))..... (-22.24 = -22.20 + -0.04)



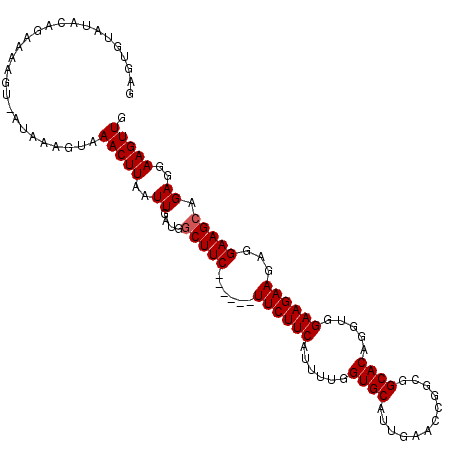
| Location | 14,159,113 – 14,159,219 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 87.69 |
| Mean single sequence MFE | -32.28 |
| Consensus MFE | -19.08 |
| Energy contribution | -19.28 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14159113 106 - 23771897 GAGUGUAUACAGAAAAGU-AUGGAGUAAACUUAAUUGAUGGCUUC------UUCUUCAUUUUGGUGCAUUGAACCGGCGGCACAGGUGGAAGAAGAGGAAGCAGAGGAAGUUG .(.(.(((((......))-))).).).(((((..((....(((((------((((((.(((..((((............))))....))).))))))))))).))..))))). ( -29.20) >DroSec_CAF1 156798 106 - 1 GCGUGUAUACAGAAAGGU-AUAAAGUGAACUUAAUUGCUGUCUUC------UUCUUCAUUUUGGUGCAUUGAACCGGCGGCACAGGUGGAAGAAGAGGAAGCAGAGGAAGUUG ((...(((((......))-)))..)).(((((..(((((.(((((------((((((......((((............)))).....))))))))))))))))...))))). ( -33.00) >DroSim_CAF1 159893 106 - 1 GCGUGUAUACAGAAAGGU-AUAAAGUAAACUUUAUUGAUGGCUUC------UUCUUCAUUUUGGUGCAUUGAACCGGUGGCACAGGUGGAAGAAGAGGAAGCAGAGGAAGUUG ((...(((((......))-)))..)).((((((.((....(((((------((((((.(((..((((((((...)))).))))....))).))))))))))).)).)))))). ( -31.20) >DroEre_CAF1 157656 109 - 1 GAGUGUAAAAC-AGUUGUAGUAAAGGAAACUUAAUUGAUGGCUUCUUC---UUCUUCAUUUCGGUGCAUUGAACCGGCGGCACAGGUAGAAGAAGAGGAAGCAGAGGAAGUUG ..........(-(....((((.(((....))).)))).))((((((((---((((((...(((((.......)))))..((....)).))))))))))))))........... ( -33.20) >DroYak_CAF1 164343 112 - 1 UAGUGUAUACAAAGUAGU-GUAAAGAAAACUUAAUUGGUGGCUUCUUCUUCUUCUUCAUUUUGGUGCAUUGAACCGGCGGCACAGGUGGAAGAAGAGGAAGCAGAGGAAGUUG .....(((((......))-))).....(((((..((....(((((((((((((((...((.((.(((.........))).)).))..))))))))))))))).))..))))). ( -34.80) >consensus GAGUGUAUACAGAAAAGU_AUAAAGUAAACUUAAUUGAUGGCUUC______UUCUUCAUUUUGGUGCAUUGAACCGGCGGCACAGGUGGAAGAAGAGGAAGCAGAGGAAGUUG ...........................(((((..((....(((((......((((((......((((............)))).....))))))...))))).))..))))). (-19.08 = -19.28 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:10 2006