
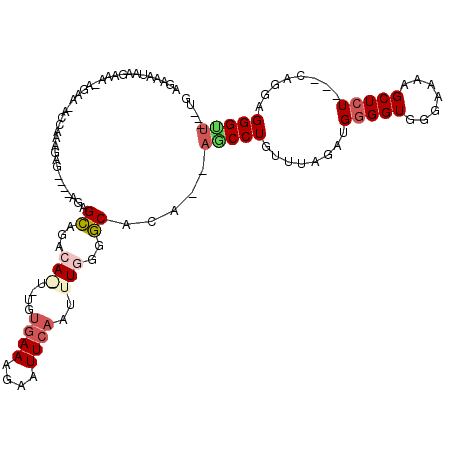
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,158,832 – 14,158,945 |
| Length | 113 |
| Max. P | 0.986857 |

| Location | 14,158,832 – 14,158,945 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.77 |
| Mean single sequence MFE | -21.44 |
| Consensus MFE | -8.21 |
| Energy contribution | -9.23 |
| Covariance contribution | 1.02 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14158832 113 + 23771897 AGAAAUAAGAUA-AGAA-ACCAAAGAGAGAGAGAGCAGACAAUAUGUGAAAGAAUUCAAUUUGGGGCACG--AACCUGUUUAGAUGGGGUGGGAAAAGCUCU---CAGGAGGGUUGAAUG ............-...(-(((.....(((((...((...(((....((((....))))..)))..))...--..((..(((.....)))..)).....))))---).....))))..... ( -20.90) >DroSec_CAF1 156527 106 + 1 AAAAAUAAGAAA-AGAA-ACCAAAGAG----AGAGCAGACAGUCUGUGAAAGAAUUCAAUUUGGGGCACA--AUCCUGUUAAGAUGGGGUGGGAAAAGCUCU---CAGGAGGGGU---UG ............-...(-(((.....(----(((((.....((((.((((....)))).....)))).(.--(((((((....))))))).).....)))))---)......)))---). ( -23.40) >DroSim_CAF1 159622 106 + 1 AAAAAUAAGAAA-AGAA-ACCAAAGAG----AGAGCAGACAGUCUGUGAAAGAAUUCAAUUUGGGGCACA--AGCCUGUUAAGAUGGGGUGGGAAAAGCUCU---CAGGAGGGGU---UG ............-...(-(((.....(----(((((...((.((((((((....)))......((((...--.))))......))))).))......)))))---)......)))---). ( -22.70) >DroEre_CAF1 157391 100 + 1 AGAAAUACGAAAUAGUAGCCCAAAUGA-----GGGGAG--------AAAAGGAAUUCAAUU-GAGCCAGAAAAGCCUGCUUAGAUGGGGUGAGAAAAGCUGU---AAGAAGGGUU---GG ...............((((((......-----......--------.........(((.((-((((.((......)))))))).)))(((.......)))..---.....)))))---). ( -15.90) >DroYak_CAF1 164071 98 + 1 AGAACUGAGAAUGAG---UCCAAAUGA-----AGGGAG--------UGAAGGCAUUCAAUU-GUGCCAGU--AGCCUGUUUAGAUGGGGUGGGAAAAGCUCUCAGAAGAAGGGUG---GA ....((((((.(.(.---((((..(((-----(.((.(--------(...(((((......-)))))...--.)))).))))..)))).).).......))))))..........---.. ( -24.30) >consensus AGAAAUAAGAAA_AGAA_ACCAAAGAG____AGAGCAGACA_U_UGUGAAAGAAUUCAAUUUGGGGCACA__AGCCUGUUUAGAUGGGGUGGGAAAAGCUCU___CAGGAGGGUU___UG ..................................((...(((....((((....))))..)))..)).....(((((........(((((.......)))))........)))))..... ( -8.21 = -9.23 + 1.02)


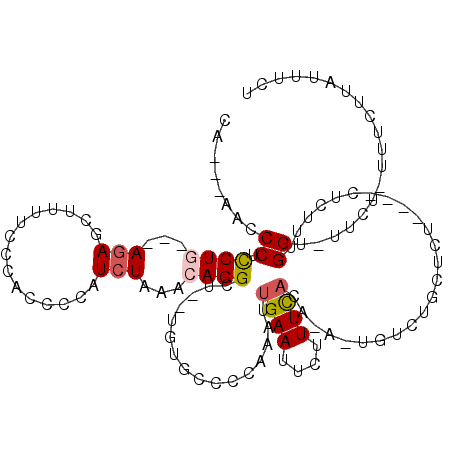
| Location | 14,158,832 – 14,158,945 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.77 |
| Mean single sequence MFE | -15.31 |
| Consensus MFE | -2.52 |
| Energy contribution | -2.92 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.16 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14158832 113 - 23771897 CAUUCAACCCUCCUG---AGAGCUUUUCCCACCCCAUCUAAACAGGUU--CGUGCCCCAAAUUGAAUUCUUUCACAUAUUGUCUGCUCUCUCUCUCUUUGGU-UUCU-UAUCUUAUUUCU .....((((.....(---(((((.....................(((.--...)))......((((....))))..........)))))).........)))-)...-............ ( -14.24) >DroSec_CAF1 156527 106 - 1 CA---ACCCCUCCUG---AGAGCUUUUCCCACCCCAUCUUAACAGGAU--UGUGCCCCAAAUUGAAUUCUUUCACAGACUGUCUGCUCU----CUCUUUGGU-UUCU-UUUCUUAUUUUU .(---(((......(---(((((......(((...((((.....))))--.)))........((((....))))..........)))))----).....)))-)...-............ ( -16.20) >DroSim_CAF1 159622 106 - 1 CA---ACCCCUCCUG---AGAGCUUUUCCCACCCCAUCUUAACAGGCU--UGUGCCCCAAAUUGAAUUCUUUCACAGACUGUCUGCUCU----CUCUUUGGU-UUCU-UUUCUUAUUUUU .(---(((......(---(((((.....................(((.--...)))......((((....))))..........)))))----).....)))-)...-............ ( -18.10) >DroEre_CAF1 157391 100 - 1 CC---AACCCUUCUU---ACAGCUUUUCUCACCCCAUCUAAGCAGGCUUUUCUGGCUC-AAUUGAAUUCCUUUU--------CUCCCC-----UCAUUUGGGCUACUAUUUCGUAUUUCU ..---.........(---((.((((..............)))).........((((((-((.(((.........--------......-----))).)))))))).......)))..... ( -13.50) >DroYak_CAF1 164071 98 - 1 UC---CACCCUUCUUCUGAGAGCUUUUCCCACCCCAUCUAAACAGGCU--ACUGGCAC-AAUUGAAUGCCUUCA--------CUCCCU-----UCAUUUGGA---CUCAUUCUCAGUUCU ..---..........(((((((....(((..............((...--.))((((.-.......))))....--------......-----......)))---....))))))).... ( -14.50) >consensus CA___AACCCUCCUG___AGAGCUUUUCCCACCCCAUCUAAACAGGCU__UGUGCCCCAAAUUGAAUUCUUUCACA_A_UGUCUGCUCU____CUCUUUGGU_UUCU_UUUCUUAUUUCU ........((.((((...(((...............)))...))))................((((....)))).........................))................... ( -2.52 = -2.92 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:07 2006