
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,154,850 – 14,154,952 |
| Length | 102 |
| Max. P | 0.967326 |

| Location | 14,154,850 – 14,154,952 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.01 |
| Mean single sequence MFE | -29.52 |
| Consensus MFE | -16.24 |
| Energy contribution | -18.22 |
| Covariance contribution | 1.97 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14154850 102 + 23771897 UGCCAAUGCAACAGCUGAACUUGAGCUCCUUUGCUUUGUUUACGCUGUGUUGCUACGACAUCG--UACCUGUUGCACC---UGUUCAACAAGUCUCAUUGGCCUCU---U .(((((((((((((((((((..((((......)))).))))).)))))..)))((((....))--))..(((((....---....)))))......))))))....---. ( -30.00) >DroGri_CAF1 148188 105 + 1 UGCCAAUGCAACAGCUGAACUUGAGCUCAAUUU-AUUGUUUACAAGCCGUUGCCAGG-CAUCA--AACUUGUUGCAUUUUCAGUUCAACAAGUGU-CUCGUCCUUAUCCU ((((...(((((.(((.....(((((.......-...)))))..))).)))))..))-))...--.((((((((.((.....)).))))))))..-.............. ( -25.50) >DroSec_CAF1 152719 104 + 1 UGCCAAUGCAACAGCUGAACUUGAGCUCCUUUGCUUUGUUUACGCUGUGUUGCUACGACAUCGUGUACCUGUUGCACC---UGUUCAACAAGUCUCAUUGGCCUCU---U .(((((((..((((((((((..((((......)))).))))).)))))((((....((((..(((((.....))))).---))))))))......)))))))....---. ( -33.10) >DroSim_CAF1 155655 105 + 1 UGCCAAUGCAACAGUUGAACUUGAGCUCCUUUGCUUUGUUUACGCUGUGUUGUUACGACAUCGUGUACCUGUUGCACGA--UGUUCAACAAGUGGGGGGGGGAUAA---A ..((..(((.((((((((((..((((......)))).))))).))))).(((((..(((((((((((.....)))))))--)))).))))))))..))........---. ( -34.30) >DroEre_CAF1 153350 102 + 1 UGCCAAUGCAACAGCUGAACUUGGGCUCAUUUGCAUUGUUUACGCUGUGUUGCUACGACAUCG--UACCUGUUGCACC---UGUUCAACAAGUCUCGUUGGCCUCU---U .(((((((((((((((((((....((......))...))))).)))))..)))((((....))--))..(((((....---....)))))......))))))....---. ( -28.00) >DroAna_CAF1 150610 99 + 1 UGCCAAUGCAACAGCUGAACUUGAGCUCCUUUGGUCUGUUUCCUCUGCGUUGCCAGGACAUUG--CACCUGUUGCUU----GGAUCAACAAGUC-CACUGUCCAUU---- ((.((((((((((((..((..........))..).))))......))))))).))(((((.((--.((.(((((...----....))))).)).-)).)))))...---- ( -26.20) >consensus UGCCAAUGCAACAGCUGAACUUGAGCUCCUUUGCUUUGUUUACGCUGUGUUGCUACGACAUCG__UACCUGUUGCACC___UGUUCAACAAGUCUCAUUGGCCUCU___U .(((((((((((((((((((..((((......)))).))))).)))..))))).............((.(((((.((.....)).))))).))...))))))........ (-16.24 = -18.22 + 1.97)


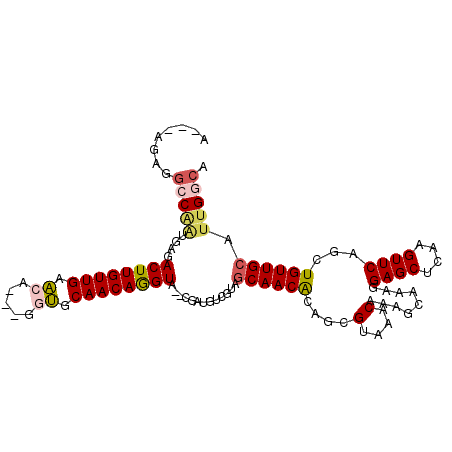
| Location | 14,154,850 – 14,154,952 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.01 |
| Mean single sequence MFE | -31.28 |
| Consensus MFE | -20.14 |
| Energy contribution | -20.62 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14154850 102 - 23771897 A---AGAGGCCAAUGAGACUUGUUGAACA---GGUGCAACAGGUA--CGAUGUCGUAGCAACACAGCGUAAACAAAGCAAAGGAGCUCAAGUUCAGCUGUUGCAUUGGCA .---....(((((((..((..(((((((.---.(.((..(.(.((--((....)))).)......((.........))...)..)).)..))))))).))..))))))). ( -34.00) >DroGri_CAF1 148188 105 - 1 AGGAUAAGGACGAG-ACACUUGUUGAACUGAAAAUGCAACAAGUU--UGAUG-CCUGGCAACGGCUUGUAAACAAU-AAAUUGAGCUCAAGUUCAGCUGUUGCAUUGGCA ..........(((.-..((((((((...(....)..)))))))))--)).((-((..(((((((((((....))).-....(((((....)))))))))))))...)))) ( -32.10) >DroSec_CAF1 152719 104 - 1 A---AGAGGCCAAUGAGACUUGUUGAACA---GGUGCAACAGGUACACGAUGUCGUAGCAACACAGCGUAAACAAAGCAAAGGAGCUCAAGUUCAGCUGUUGCAUUGGCA .---....(((((((..((..(((((((.---.((((.....))))(((.(((.((....))))).)))......(((......)))...))))))).))..))))))). ( -31.90) >DroSim_CAF1 155655 105 - 1 U---UUAUCCCCCCCCCACUUGUUGAACA--UCGUGCAACAGGUACACGAUGUCGUAACAACACAGCGUAAACAAAGCAAAGGAGCUCAAGUUCAACUGUUGCAUUGGCA .---................(((..((((--(((((.........)))))))).((((((.....((.........))....((((....))))...)))))).)..))) ( -23.20) >DroEre_CAF1 153350 102 - 1 A---AGAGGCCAACGAGACUUGUUGAACA---GGUGCAACAGGUA--CGAUGUCGUAGCAACACAGCGUAAACAAUGCAAAUGAGCCCAAGUUCAGCUGUUGCAUUGGCA .---....(((((.(..((..(((((((.---((.((....(.((--((....)))).)......((((.....))))......))))..))))))).))..).))))). ( -31.70) >DroAna_CAF1 150610 99 - 1 ----AAUGGACAGUG-GACUUGUUGAUCC----AAGCAACAGGUG--CAAUGUCCUGGCAACGCAGAGGAAACAGACCAAAGGAGCUCAAGUUCAGCUGUUGCAUUGGCA ----...(((((.((-.((((((((....----...)))))))).--)).)))))..(((((((...(....).........((((....)))).)).)))))....... ( -34.80) >consensus A___AGAGGCCAAUGAGACUUGUUGAACA___GGUGCAACAGGUA__CGAUGUCGUAGCAACACAGCGUAAACAAAGCAAAGGAGCUCAAGUUCAGCUGUUGCAUUGGCA ........(((((....((((((((.((.....)).)))))))).............((((((....(....).........((((....))))...)))))).))))). (-20.14 = -20.62 + 0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:02 2006