
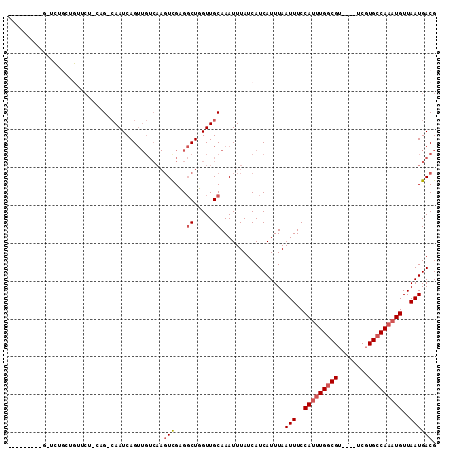
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,137,184 – 14,137,275 |
| Length | 91 |
| Max. P | 0.562896 |

| Location | 14,137,184 – 14,137,275 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 78.24 |
| Mean single sequence MFE | -24.43 |
| Consensus MFE | -12.82 |
| Energy contribution | -13.43 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14137184 91 + 23771897 ---------G--------UUCUCCUG-CAAUCAGUUGUCAAGUUGCGGCUGGUUGCAAAUUUAUCAUCAUUUAAUUUCCAUUUGGCGU----UCGUGCCAAAUGUUAAUGACG ---------(--------((....((-(((((((((((......)))))))))))))................(((..(((((((((.----...)))))))))..)))))). ( -30.40) >DroVir_CAF1 135029 83 + 1 -----------AUUGCCGC----ACGCAAAUCAG--------UCGAGUCUUGUUGCAAAUUUAUCAUCAUUUAAUUUCCAUUUGGCG-----UCGUGCCA--UGUUAAUGACG -----------...(((((----(.((((.((..--------..))...)))))))....(((........))).........)))(-----((((....--.....))))). ( -13.00) >DroPse_CAF1 151559 111 + 1 UUUCCACUCAUUCUGCUGUGCU-CAG-CAAUCAGUUGUCAAGUCGAGGCUUGUUGCAAAUUUAUCAUCAUUUAAUUUCCAUUUGGCGUUCGUUCGUGCCAAAUGUUAAUGACG .............(((((....-)))-))....((...(((((....)))))..)).................(((..(((((((((........)))))))))..))).... ( -25.00) >DroYak_CAF1 141526 99 + 1 ---------GUUCUGCUGUUCUCCUG-CAAUCAGUUGUCAAGUUGCGGCUGGUUGCAAAUUUAUCAUCAUUUAAUUUCCAUUUGGCGU----UCGUGCCAAAUGUUAAUGACG ---------........(((....((-(((((((((((......)))))))))))))................(((..(((((((((.----...)))))))))..)))))). ( -30.40) >DroAna_CAF1 134666 89 + 1 ---------G--------UUC--AAG-CAAUCAGUUGUCAAGUCGCGGCUGGUUGCAAAUUUAUCAUCAUUUAAUUUCCAUUUGCCGU----UCGUGCCAAAUGUUAAUGACG ---------(--------((.--..(-(((((((((((......)))))))))))).................(((..((((((.((.----...)).))))))..)))))). ( -22.80) >DroPer_CAF1 152858 111 + 1 UUUCCACUCAUUCUGCUGUGCU-CAG-CAAUCAGUUGUCAAGUCGAGGCUUGUUGCAAAUUUAUCAUCAUUUAAUUUCCAUUUGGCGUUCGUUCGUGCCAAAUGUUAAUGACG .............(((((....-)))-))....((...(((((....)))))..)).................(((..(((((((((........)))))))))..))).... ( -25.00) >consensus _________G_UCUGCUGUUCU_CAG_CAAUCAGUUGUCAAGUCGAGGCUGGUUGCAAAUUUAUCAUCAUUUAAUUUCCAUUUGGCGU____UCGUGCCAAAUGUUAAUGACG .........................................(((...((.....)).................(((..(((((((((........)))))))))..)))))). (-12.82 = -13.43 + 0.61)


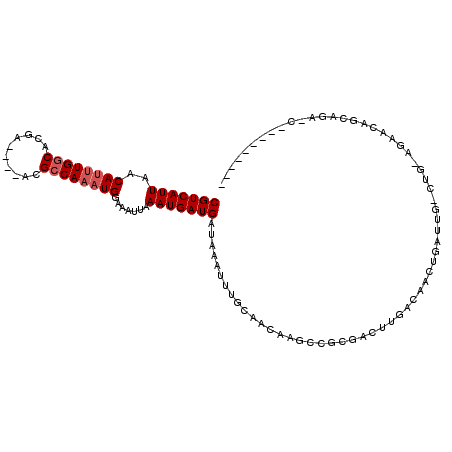
| Location | 14,137,184 – 14,137,275 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 78.24 |
| Mean single sequence MFE | -22.34 |
| Consensus MFE | -12.60 |
| Energy contribution | -13.10 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.546828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14137184 91 - 23771897 CGUCAUUAACAUUUGGCACGA----ACGCCAAAUGGAAAUUAAAUGAUGAUAAAUUUGCAACCAGCCGCAACUUGACAACUGAUUG-CAGGAGAA--------C--------- (((((((..((((((((....----..)))))))).......))))))).....(((((((.(((...(.....)....))).)))-))))....--------.--------- ( -23.70) >DroVir_CAF1 135029 83 - 1 CGUCAUUAACA--UGGCACGA-----CGCCAAAUGGAAAUUAAAUGAUGAUAAAUUUGCAACAAGACUCGA--------CUGAUUUGCGU----GCGGCAAU----------- (((((((....--((((....-----.))))(((....))).)))))))......((((..((.((.((..--------..)).))...)----)..)))).----------- ( -16.50) >DroPse_CAF1 151559 111 - 1 CGUCAUUAACAUUUGGCACGAACGAACGCCAAAUGGAAAUUAAAUGAUGAUAAAUUUGCAACAAGCCUCGACUUGACAACUGAUUG-CUG-AGCACAGCAGAAUGAGUGGAAA (((((((..((((((((..........)))))))).......))))))).....((..(..((((......))))........(((-(((-....)))))).....)..)).. ( -26.60) >DroYak_CAF1 141526 99 - 1 CGUCAUUAACAUUUGGCACGA----ACGCCAAAUGGAAAUUAAAUGAUGAUAAAUUUGCAACCAGCCGCAACUUGACAACUGAUUG-CAGGAGAACAGCAGAAC--------- (((((((..((((((((....----..)))))))).......))))))).....(((((......(((((((.........).)))-).))......)))))..--------- ( -24.50) >DroAna_CAF1 134666 89 - 1 CGUCAUUAACAUUUGGCACGA----ACGGCAAAUGGAAAUUAAAUGAUGAUAAAUUUGCAACCAGCCGCGACUUGACAACUGAUUG-CUU--GAA--------C--------- (((((((..((((((.(....----..).)))))).......)))))))........((((.(((..............))).)))-)..--...--------.--------- ( -16.14) >DroPer_CAF1 152858 111 - 1 CGUCAUUAACAUUUGGCACGAACGAACGCCAAAUGGAAAUUAAAUGAUGAUAAAUUUGCAACAAGCCUCGACUUGACAACUGAUUG-CUG-AGCACAGCAGAAUGAGUGGAAA (((((((..((((((((..........)))))))).......))))))).....((..(..((((......))))........(((-(((-....)))))).....)..)).. ( -26.60) >consensus CGUCAUUAACAUUUGGCACGA____ACGCCAAAUGGAAAUUAAAUGAUGAUAAAUUUGCAACAAGCCGCGACUUGACAACUGAUUG_CUG_AGAACAGCAGA_C_________ (((((((..((((((((..........)))))))).......)))))))................................................................ (-12.60 = -13.10 + 0.50)

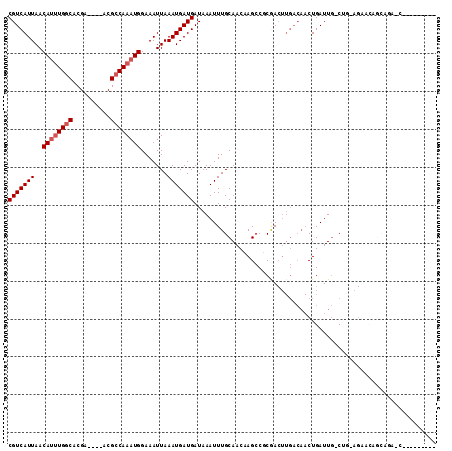
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:56 2006