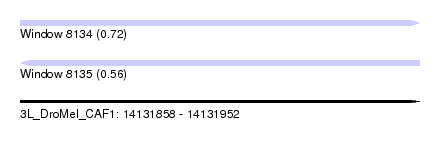
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,131,858 – 14,131,952 |
| Length | 94 |
| Max. P | 0.724856 |

| Location | 14,131,858 – 14,131,952 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.63 |
| Mean single sequence MFE | -29.01 |
| Consensus MFE | -22.76 |
| Energy contribution | -22.23 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
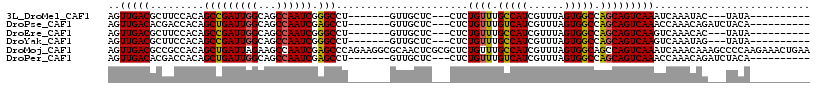
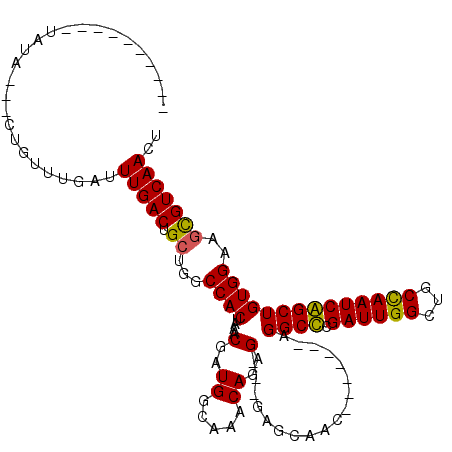
>3L_DroMel_CAF1 14131858 94 + 23771897 AGUUGACGCUUCCACAGCCGAUUGGCAGCCAAUCGGGCCU-------GUUGCUC---CUCUGUUUGCCAUCGUUUAGUGGCCAGCAGUCAAAUCAAAUAC---UAUA---------- ..(((((((....((((((((((((...))))))))..))-------)).))..---...((((.(((((......))))).))))))))).........---....---------- ( -29.90) >DroPse_CAF1 146574 97 + 1 AGUUGACACGACCACAGCUGAUUGGCAGCCAAUCGAGCCU-------GUUGCUC---CUCUGUUUGUCAUCGUUUAGUGGCCAGCAGUCAAACCAAACAGAUCUACA---------- .((.((..........(((((((((...)))))).)))((-------(((....---..(((((.(((((......))))).)))))........))))).)).)).---------- ( -24.44) >DroEre_CAF1 128866 94 + 1 AGUUGACGCUUCCACAGCCGAUUGGCAGCCAAUCGGGCCU-------GUUGCUC---CUCUGUUUGCCAUCGUUUAGUGGCCAGCAGUCAAGUCAAACAC---UAUA---------- ..(((((((....((((((((((((...))))))))..))-------)).))..---..(((((.(((((......))))).)))))....)))))....---....---------- ( -32.30) >DroYak_CAF1 135637 94 + 1 AGUUGACGCUUCCACAGCCGAUUGGCAGCCAAUCGGGCCU-------GUUGCUC---CUCUGUUUGCCAUCGUUUAGUGGCCAGCAGUCAAGUCAAAUAG---UAUA---------- ..(((((((....((((((((((((...))))))))..))-------)).))..---..(((((.(((((......))))).)))))....)))))....---....---------- ( -32.30) >DroMoj_CAF1 143354 117 + 1 AGUUGACGCCGCCACAGCUGAUUAGAAGCCAAUCGAGCCCAGAAGGCGCAACUCGCGCUCUGUUUGCCAUCGUUUAGUGGCAGCCAGUCAAAUCAAACAAAGCCCCAAGAAACUGAA ..(((((((.(((((.(((.......)))....(((...((((.(((((.....)))))...))))...)))....))))).))..))))).......................... ( -30.70) >DroPer_CAF1 147906 97 + 1 AGUUGACACGACCACAGCUGAUUGGCAGCCAAUCGAGCCU-------GUUGCUC---CUCUGUUUGUCAUCGUUUAGUGGCCAGCAGUCAAACCAAACAGAUCUACA---------- .((.((..........(((((((((...)))))).)))((-------(((....---..(((((.(((((......))))).)))))........))))).)).)).---------- ( -24.44) >consensus AGUUGACGCUUCCACAGCCGAUUGGCAGCCAAUCGAGCCU_______GUUGCUC___CUCUGUUUGCCAUCGUUUAGUGGCCAGCAGUCAAAUCAAACAG___UACA__________ ..(((((.........(((((((((...)))))).)))......................((((.(((((......))))).))))))))).......................... (-22.76 = -22.23 + -0.53)
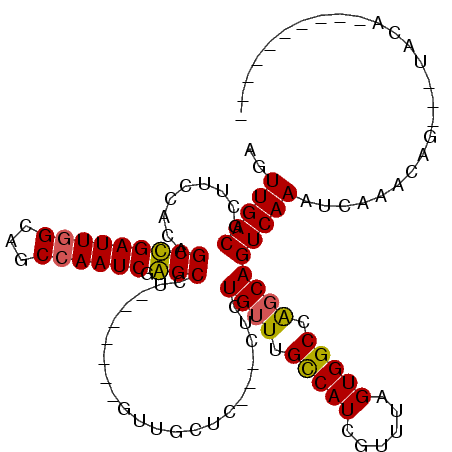
| Location | 14,131,858 – 14,131,952 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.63 |
| Mean single sequence MFE | -32.12 |
| Consensus MFE | -21.68 |
| Energy contribution | -20.98 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14131858 94 - 23771897 ----------UAUA---GUAUUUGAUUUGACUGCUGGCCACUAAACGAUGGCAAACAGAG---GAGCAAC-------AGGCCCGAUUGGCUGCCAAUCGGCUGUGGAAGCGUCAACU ----------....---.........(((((.(((..((((....((((((((..(((.(---(.((...-------..))))..)))..)))).))))...)))).)))))))).. ( -33.30) >DroPse_CAF1 146574 97 - 1 ----------UGUAGAUCUGUUUGGUUUGACUGCUGGCCACUAAACGAUGACAAACAGAG---GAGCAAC-------AGGCUCGAUUGGCUGCCAAUCAGCUGUGGUCGUGUCAACU ----------.((.(((.((((((((....(....)...))))))))(((((..((((..---((((...-------..))))((((((...))))))..)))).)))))))).)). ( -26.60) >DroEre_CAF1 128866 94 - 1 ----------UAUA---GUGUUUGACUUGACUGCUGGCCACUAAACGAUGGCAAACAGAG---GAGCAAC-------AGGCCCGAUUGGCUGCCAAUCGGCUGUGGAAGCGUCAACU ----------....---.........(((((.(((..((((....((((((((..(((.(---(.((...-------..))))..)))..)))).))))...)))).)))))))).. ( -33.20) >DroYak_CAF1 135637 94 - 1 ----------UAUA---CUAUUUGACUUGACUGCUGGCCACUAAACGAUGGCAAACAGAG---GAGCAAC-------AGGCCCGAUUGGCUGCCAAUCGGCUGUGGAAGCGUCAACU ----------....---.........(((((.(((..((((....((((((((..(((.(---(.((...-------..))))..)))..)))).))))...)))).)))))))).. ( -33.20) >DroMoj_CAF1 143354 117 - 1 UUCAGUUUCUUGGGGCUUUGUUUGAUUUGACUGGCUGCCACUAAACGAUGGCAAACAGAGCGCGAGUUGCGCCUUCUGGGCUCGAUUGGCUUCUAAUCAGCUGUGGCGGCGUCAACU ............(.((((((((((..(((..(((......)))..)))...)))))))))).).(((((((((.((..((((.((((((...))))))))))..)).)))).))))) ( -39.80) >DroPer_CAF1 147906 97 - 1 ----------UGUAGAUCUGUUUGGUUUGACUGCUGGCCACUAAACGAUGACAAACAGAG---GAGCAAC-------AGGCUCGAUUGGCUGCCAAUCAGCUGUGGUCGUGUCAACU ----------.((.(((.((((((((....(....)...))))))))(((((..((((..---((((...-------..))))((((((...))))))..)))).)))))))).)). ( -26.60) >consensus __________UAUA___CUGUUUGAUUUGACUGCUGGCCACUAAACGAUGGCAAACAGAG___GAGCAAC_______AGGCCCGAUUGGCUGCCAAUCAGCUGUGGAAGCGUCAACU ..........................(((((.((...((((....(..((.....))..)..................((((.((((((...))))))))))))))..))))))).. (-21.68 = -20.98 + -0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:52 2006