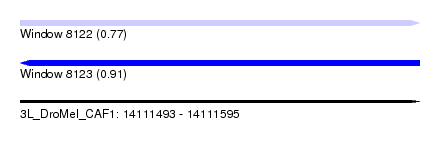
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,111,493 – 14,111,595 |
| Length | 102 |
| Max. P | 0.910584 |

| Location | 14,111,493 – 14,111,595 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 77.54 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -18.13 |
| Energy contribution | -18.30 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14111493 102 + 23771897 ------GU--UGGAUGAAAAAAUGGAAAUGUUGCUGCUCUAUUGAUUAUUGGAUAUUGAUUGCUG-GUGCUGCCUUUUAAUUCGAGGCAGGCAGCAACAAGCAGCCGCCAG ------..--(((.((.....((....))...(((((((((........))))..(((.((((((-...(((((((.......))))))).))))))))))))))))))). ( -29.90) >DroVir_CAF1 110757 93 + 1 -----------GGAAAUGAAAAUGAAAAUGUUGUUGCUCUAUUGAUUAUUGCAUAUUGAUUGCUGUGUGGCUCUUUUUUA-UUGAGGCGAGCAGCGACAAAUAAA------ -----------.................(((((((((((.......(((.(((.......))).))).(.(((.......-..))).))))))))))))......------ ( -23.80) >DroPse_CAF1 124922 89 + 1 ------GU--UGG-----AAAAUGGAAAUGUUG---CUCUAUUGAUUAUUGGAUAUUGAUUGCUGUGCGCUGCCUUUUAAUGCGAGGCGAGCAGCAACAAGAAAC------ ------..--...-----..........(((((---(((....(((((........)))))(((...(((...........))).))))))))))).........------ ( -20.50) >DroEre_CAF1 108803 107 + 1 CAU---GUGCUGGAUGGAAAAAUGGAAAUGUUGCUGCUCUAUUGAUUAUUGGAUAUUGAUUGCUG-GUGCUGCCUUUUAAUUCGAGGCAGGCAGCAACAAGCAGCCGAAAG ..(---(.((((..(....)........(((((((((.(((..(((((........)))))..))-)..(((((((.......))))))))))))))))..)))))).... ( -31.50) >DroAna_CAF1 111072 103 + 1 GAUGGUAU--CGGCUG-GAAAAUGGAAAUGUUGCUGCUCUAUUGAUUAUUGGAUAUUGAUUGCUG-GGGCUGCCUUUUAAUUCGAGGCAGGCAGCAACAAGC----GACAG ........--..(((.-....((....))((((((((((((..(((((........)))))..))-)).(((((((.......))))))))))))))).)))----..... ( -29.80) >DroPer_CAF1 125939 89 + 1 ------GU--UGG-----AAAAUGGAAAUGUUG---CUCUAUUGAUUAUUGGAUAUUGAUUGCUGUGCGCUGCCUUUUAAUGCGAGGCGAGCAGCAACAAGAAAC------ ------..--...-----..........(((((---(((....(((((........)))))(((...(((...........))).))))))))))).........------ ( -20.50) >consensus ______GU__UGGAUG__AAAAUGGAAAUGUUGCUGCUCUAUUGAUUAUUGGAUAUUGAUUGCUG_GUGCUGCCUUUUAAUUCGAGGCAAGCAGCAACAAGCAAC______ ............................((((((((((.....(((((........))))).........((((((.......))))))))))))))))............ (-18.13 = -18.30 + 0.17)

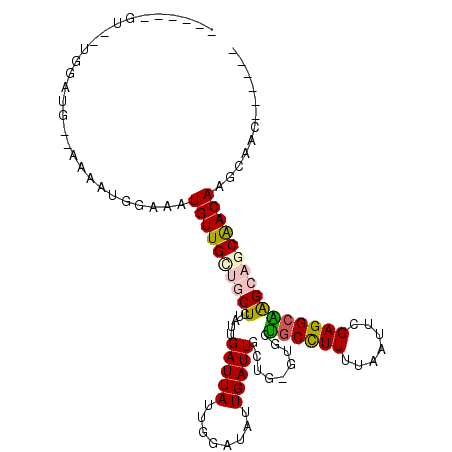
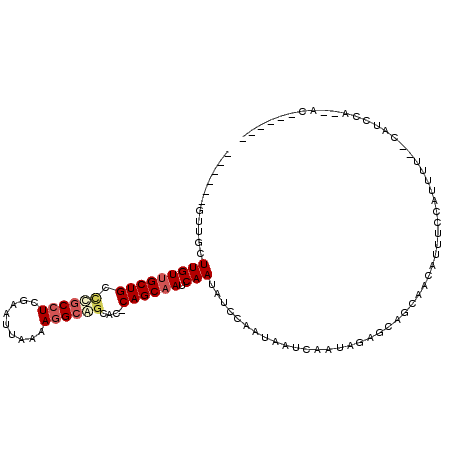
| Location | 14,111,493 – 14,111,595 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.54 |
| Mean single sequence MFE | -20.40 |
| Consensus MFE | -10.60 |
| Energy contribution | -11.27 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14111493 102 - 23771897 CUGGCGGCUGCUUGUUGCUGCCUGCCUCGAAUUAAAAGGCAGCAC-CAGCAAUCAAUAUCCAAUAAUCAAUAGAGCAGCAACAUUUCCAUUUUUUCAUCCA--AC------ ...(..((((((((((((((.((((((.........))))))...-)))))))...................)))))))..)...................--..------ ( -26.30) >DroVir_CAF1 110757 93 - 1 ------UUUAUUUGUCGCUGCUCGCCUCAA-UAAAAAAGAGCCACACAGCAAUCAAUAUGCAAUAAUCAAUAGAGCAACAACAUUUUCAUUUUCAUUUCC----------- ------......(((.(.((((((.(((..-.......))).).....(((.......)))...........))))).).))).................----------- ( -12.70) >DroPse_CAF1 124922 89 - 1 ------GUUUCUUGUUGCUGCUCGCCUCGCAUUAAAAGGCAGCGCACAGCAAUCAAUAUCCAAUAAUCAAUAGAG---CAACAUUUCCAUUUU-----CCA--AC------ ------......((((((((((.((((.........)))))))((...)).......................))---)))))..........-----...--..------ ( -16.90) >DroEre_CAF1 108803 107 - 1 CUUUCGGCUGCUUGUUGCUGCCUGCCUCGAAUUAAAAGGCAGCAC-CAGCAAUCAAUAUCCAAUAAUCAAUAGAGCAGCAACAUUUCCAUUUUUCCAUCCAGCAC---AUG ......((((((((((((((.((((((.........))))))...-)))))))...................)))))))..........................---... ( -25.60) >DroAna_CAF1 111072 103 - 1 CUGUC----GCUUGUUGCUGCCUGCCUCGAAUUAAAAGGCAGCCC-CAGCAAUCAAUAUCCAAUAAUCAAUAGAGCAGCAACAUUUCCAUUUUC-CAGCCG--AUACCAUC .((((----((((((((((((((((((.........))))))...-....................((....)))))))))))...........-.)).))--)))..... ( -24.00) >DroPer_CAF1 125939 89 - 1 ------GUUUCUUGUUGCUGCUCGCCUCGCAUUAAAAGGCAGCGCACAGCAAUCAAUAUCCAAUAAUCAAUAGAG---CAACAUUUCCAUUUU-----CCA--AC------ ------......((((((((((.((((.........)))))))((...)).......................))---)))))..........-----...--..------ ( -16.90) >consensus ______GUUGCUUGUUGCUGCCCGCCUCGAAUUAAAAGGCAGCAC_CAGCAAUCAAUAUCCAAUAAUCAAUAGAGCAGCAACAUUUCCAUUUU__CAUCCA__AC______ ...........(((((((((.((((((.........))))))....)))))).)))....................................................... (-10.60 = -11.27 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:40 2006