
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,094,350 – 14,094,445 |
| Length | 95 |
| Max. P | 0.576730 |

| Location | 14,094,350 – 14,094,445 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 82.78 |
| Mean single sequence MFE | -28.75 |
| Consensus MFE | -19.63 |
| Energy contribution | -19.63 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
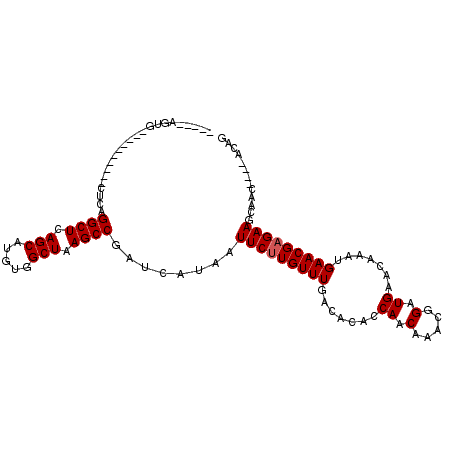
>3L_DroMel_CAF1 14094350 95 + 23771897 GUGUUGUUGCUGCUUCGCGUUCAUUUGUUCAUCCGUUUGUUGGUAUGUCAAACAAGAAUUAUGAUCGGCUUAGCCACAUGCUGAGCCCGAG----------AUCU----- (((..((....))..)))((((.((((..((((((.....))).))).))))...))))...((((((((((((.....))))))))...)----------))).----- ( -24.30) >DroVir_CAF1 93091 91 + 1 CUGU----CUUGCUUCUCGUUCAUUUGUUCAUCCGUUUGUUGCAGUGUCAAACAAGAAUUAUGAUCGGCUUAGCCACAUGCUGAGCCUGUG----------CAUG----- ....----..(((.....(.((((..((((....(((((.........)))))..)))).)))).)((((((((.....))))))))...)----------))..----- ( -22.50) >DroPse_CAF1 105479 104 + 1 UUGUUGUAGCUGCUUCUCGUUCAUUUGUUCAUCCGUUUGUUGGUGUGUCAAACAAGAAUUAUGAUCGGCUUAGCCACAUGCUGAGCC-GAGCUGGAAGAGGAGCU----- ...........(((((((.((((((((((((((((.....))).)))...))))))........((((((((((.....))))))))-))..)))).))))))).----- ( -35.60) >DroGri_CAF1 89371 96 + 1 CUGU----CUUGCUUCUCGUUCAUUUGUUCAUCCGUUUGUUGGAGUGUCAAACAAGAAUUAUGAUCGGCUUAGCCACAUGCUGAGCCUGCG----------CAGAGACGA .(((----(((((.....(.((((..((((.((((.....))))((.....))..)))).)))).)((((((((.....))))))))...)----------)).))))). ( -30.30) >DroMoj_CAF1 101591 91 + 1 CUGU----CUUGCUUCUCGUUCAUUUGUUCAUCCGUUUGUUGCAGUGUCAAACAAGAAUUAUGAUCGGCUUAGCCACAUGCUGAGCCUGCG----------CAGA----- ((((----...((.....(.((((..((((....(((((.........)))))..)))).)))).)((((((((.....)))))))).)))----------))).----- ( -24.20) >DroPer_CAF1 106661 104 + 1 UUGUUGUAGCUGCUUCUCGUUCAUUUGUUCAUCCGUUUGUUGGUGUGUCAAACAAGAAUUAUGAUCGGCUUAGCCACAUGCUGAGCC-GAGCUGGAAGAGGAGCU----- ...........(((((((.((((((((((((((((.....))).)))...))))))........((((((((((.....))))))))-))..)))).))))))).----- ( -35.60) >consensus CUGU____CCUGCUUCUCGUUCAUUUGUUCAUCCGUUUGUUGGAGUGUCAAACAAGAAUUAUGAUCGGCUUAGCCACAUGCUGAGCCUGAG__________AACU_____ ..................(.((((..((((....(((((.........)))))..)))).)))).)((((((((.....))))))))....................... (-19.63 = -19.63 + 0.00)



| Location | 14,094,350 – 14,094,445 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 82.78 |
| Mean single sequence MFE | -23.81 |
| Consensus MFE | -16.17 |
| Energy contribution | -16.34 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14094350 95 - 23771897 -----AGAU----------CUCGGGCUCAGCAUGUGGCUAAGCCGAUCAUAAUUCUUGUUUGACAUACCAACAAACGGAUGAACAAAUGAACGCGAAGCAGCAACAACAC -----.(((----------(...((((.(((.....))).)))))))).......(..((((.(((.((.......)))))..))))..)..((......))........ ( -20.30) >DroVir_CAF1 93091 91 - 1 -----CAUG----------CACAGGCUCAGCAUGUGGCUAAGCCGAUCAUAAUUCUUGUUUGACACUGCAACAAACGGAUGAACAAAUGAACGAGAAGCAAG----ACAG -----..((----------(...((((.(((.....))).))))........(((((((((.......((.(.....).)).......))))))))))))..----.... ( -21.14) >DroPse_CAF1 105479 104 - 1 -----AGCUCCUCUUCCAGCUC-GGCUCAGCAUGUGGCUAAGCCGAUCAUAAUUCUUGUUUGACACACCAACAAACGGAUGAACAAAUGAACGAGAAGCAGCUACAACAA -----.(((.(((.......((-((((.(((.....))).))))))((((..(((.((((((.........))))))...)))...))))..))).)))........... ( -26.40) >DroGri_CAF1 89371 96 - 1 UCGUCUCUG----------CGCAGGCUCAGCAUGUGGCUAAGCCGAUCAUAAUUCUUGUUUGACACUCCAACAAACGGAUGAACAAAUGAACGAGAAGCAAG----ACAG ..((((.((----------(...((((.(((.....))).))))........(((((((((..((.(((.......))))).......))))))))))))))----)).. ( -27.10) >DroMoj_CAF1 101591 91 - 1 -----UCUG----------CGCAGGCUCAGCAUGUGGCUAAGCCGAUCAUAAUUCUUGUUUGACACUGCAACAAACGGAUGAACAAAUGAACGAGAAGCAAG----ACAG -----(((.----------.((.((((.(((.....))).))))........(((((((((.......((.(.....).)).......))))))))))).))----)... ( -21.54) >DroPer_CAF1 106661 104 - 1 -----AGCUCCUCUUCCAGCUC-GGCUCAGCAUGUGGCUAAGCCGAUCAUAAUUCUUGUUUGACACACCAACAAACGGAUGAACAAAUGAACGAGAAGCAGCUACAACAA -----.(((.(((.......((-((((.(((.....))).))))))((((..(((.((((((.........))))))...)))...))))..))).)))........... ( -26.40) >consensus _____AGUG__________CUCAGGCUCAGCAUGUGGCUAAGCCGAUCAUAAUUCUUGUUUGACACACCAACAAACGGAUGAACAAAUGAACGAGAAGCAAC____ACAG .......................((((.(((.....))).))))........(((((((((.......((.(.....).)).......)))))))))............. (-16.17 = -16.34 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:36 2006