
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,060,023 – 14,060,169 |
| Length | 146 |
| Max. P | 0.774495 |

| Location | 14,060,023 – 14,060,129 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 79.39 |
| Mean single sequence MFE | -29.77 |
| Consensus MFE | -19.94 |
| Energy contribution | -20.67 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
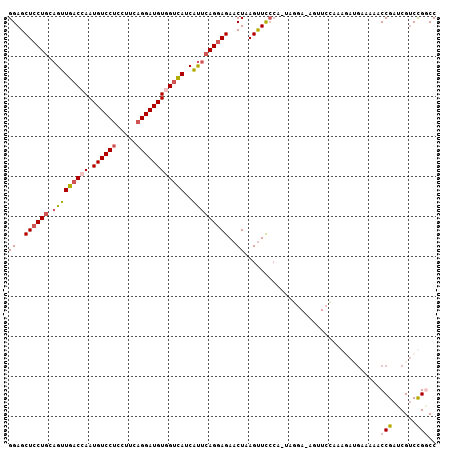
>3L_DroMel_CAF1 14060023 106 - 23771897 GGAGCUCCUGCAGUUGACCAAUGUCCUGCUUCAGGAUGUGGUCAUCAUUCAGGAGAACUAAGUUCCCAAUACGA-AGUUCCAAAUAUGUUAAACCGAUCGUCUGGCC ((((((((((....((((((.(((((((...))))))))))))).....)))))((((...)))).........-.)))))............(((......))).. ( -31.20) >DroPse_CAF1 62675 93 - 1 GGAGCUGCUCCAGCUGCCGAAUGUCCACCUGCAGGAUGUGGUUAUCGUCCAGGAGAACUAAGCUUGAA--------------CGGAUUACCAACCGACCGACCGACC (((((((...))))).))....(((..((((.(.((((....)))).).))))........(.(((..--------------(((........)))..))).)))). ( -22.00) >DroSec_CAF1 58563 107 - 1 GGAGCUCCUGCAGUUGACCAAUGUCCUCCUUCAGGAUGUUGUCAUCAUUCAGGAGAACUAAGUUCCCAUUAGGAAAGUUCCAAAGAUGUAAAACCGAUCGUCCGGCC ((((((((((....((((.((((((((.....)))))))))))).....)))))........((((.....)))).)))))............(((......))).. ( -30.60) >DroSim_CAF1 59559 106 - 1 GGAGCUCCUGCAGUUGACCAAUGUCCUCCUGCAGGAUGUGGUCAUCAUUCAGGAGAACUAAGUUCCCAUUAGGA-AGUUCCAAAGAUGUAAAACCGAUCGUCCGGCC ((((((((((..(.((((((.((((((.....)))))))))))).).....((.((((...))))))..)))).-))))))............(((......))).. ( -34.40) >DroEre_CAF1 56490 106 - 1 GGAGCUCCUGCAGUUGACCAAUGUCCUUCUUAAGGAUGUGGUCAUCACUCAGGAGAACUAAGUUCCCAAUAGAA-AGUUAUAAGCAAGAAAAUCCGAUCGUCCGACC (((.((((((.(((((((((.((((((.....))))))))))))..)))))))))((((...(((......)))-)))).............)))............ ( -29.60) >DroYak_CAF1 60049 93 - 1 GGAGCUCCUGCAGUUGACCAAUGUCCUGCUCCAGGAUGUGGUCAUCAUUCAGGAGAACUAAGUUCCCC--------------AGGAAGAAAACCCGAUCGUCUGGCC ((((((((((....((((((.(((((((...))))))))))))).....))))).......)))))((--------------(((..((........)).))))).. ( -30.81) >consensus GGAGCUCCUGCAGUUGACCAAUGUCCUCCUUCAGGAUGUGGUCAUCAUUCAGGAGAACUAAGUUCCCA_UAGGA_AGUUCCAAAGAUGAAAAACCGAUCGUCCGGCC ((..((((((.(((((((((.((((((.....))))))))))))..)))))))))..))..................................(((......))).. (-19.94 = -20.67 + 0.72)


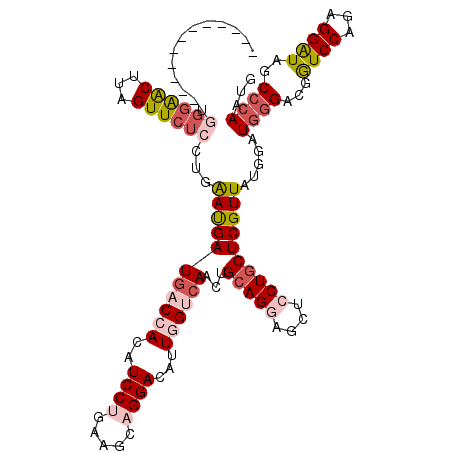
| Location | 14,060,050 – 14,060,169 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.16 |
| Mean single sequence MFE | -43.73 |
| Consensus MFE | -24.33 |
| Energy contribution | -25.92 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14060050 119 + 23771897 GAACU-UCGUAUUGGGAACUUAGUUCUCCUGAAUGAUGACCACAUCCUGAAGCAGGACAUUGGUCAACUGCAGGAGCUCCUGCUCGUUAUGGAUGGGCCGGUCCAGAGGAUAACCCAAUG .....-...(((((((......(((((((..(((((((((((..(((((...)))))...))))))...(((((....))))))))))..))).))))..((((...))))..))))))) ( -41.50) >DroPse_CAF1 62700 108 + 1 ------------UUCAAGCUUAGUUCUCCUGGACGAUAACCACAUCCUGCAGGUGGACAUUCGGCAGCUGGAGCAGCUCCUGCUCGUUAGUGAUGGGACGAUCCAGGGGGUAGCCGAGGG ------------.....(((....(((((((((((....((.((((((((((.((..(....).)).)))((((((...))))))..))).)))))).)).))))))))).)))...... ( -37.60) >DroSec_CAF1 58590 120 + 1 GAACUUUCCUAAUGGGAACUUAGUUCUCCUGAAUGAUGACAACAUCCUGAAGGAGGACAUUGGUCAACUGCAGGAGCUCCUGCUCGUUAUGGAUGGGACGGUCCAGAGGAUAGCCCAAUG ......(((((((((((.((..((((((((..(.((((....)))).)..))))))))...))))....(((((....))))))))))).)))((((...((((...))))..))))... ( -41.60) >DroSim_CAF1 59586 119 + 1 GAACU-UCCUAAUGGGAACUUAGUUCUCCUGAAUGAUGACCACAUCCUGCAGGAGGACAUUGGUCAACUGCAGGAGCUCCUGCUCGUUAUGGAUGGGACGGUCCAGAGGAUAACCCAAUG .....-(((((((((((.((..(((((((((...((((....))))...)))))))))...))))....(((((....))))))))))).)))((((...((((...))))..))))... ( -43.50) >DroYak_CAF1 60074 108 + 1 ------------GGGGAACUUAGUUCUCCUGAAUGAUGACCACAUCCUGGAGCAGGACAUUGGUCAACUGCAGGAGCUCCUGCUCGUUGCGGAUGGGCCGGUCCAGGGGAUAGCCCAAUG ------------(((((((...))))))).......((((((..(((((...)))))...)))))).((((((((((....)))).)))))).(((((..((((...)))).)))))... ( -46.60) >DroAna_CAF1 59184 107 + 1 -------------UGGGGCUUAGUUCUCCUGGACGAUGACCACAUCCUGGAGCUGGACAUUGGGCAGCUGCAGCAGGGCCUGCUCGUUGUCGAUGCGACGGUCCAGGGGAUAGCCCAGUG -------------..(((((....((((((((((((((....)))).((..(((.((((.(((((((..((......))))))))).)))).).))..)))))))))))).))))).... ( -51.60) >consensus ____________UGGGAACUUAGUUCUCCUGAAUGAUGACCACAUCCUGAAGCAGGACAUUGGUCAACUGCAGGAGCUCCUGCUCGUUAUGGAUGGGACGGUCCAGAGGAUAGCCCAAUG .............((((((...))))))...(((((((((((..((((.....))))...))))))...(((((....)))))))))).....((((...((((...))))..))))... (-24.33 = -25.92 + 1.59)


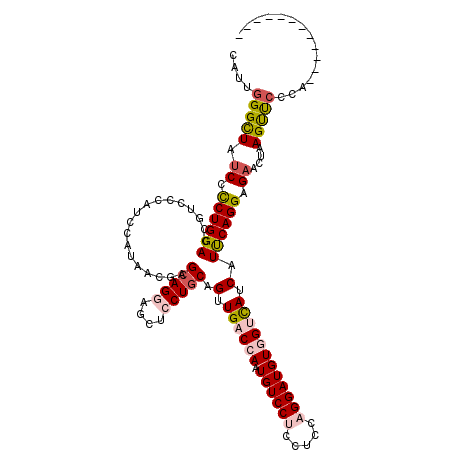
| Location | 14,060,050 – 14,060,169 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.16 |
| Mean single sequence MFE | -38.65 |
| Consensus MFE | -25.14 |
| Energy contribution | -25.78 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14060050 119 - 23771897 CAUUGGGUUAUCCUCUGGACCGGCCCAUCCAUAACGAGCAGGAGCUCCUGCAGUUGACCAAUGUCCUGCUUCAGGAUGUGGUCAUCAUUCAGGAGAACUAAGUUCCCAAUACGA-AGUUC ...((((((.(((...)))..))))))(((.......(((((....))))).(.((((((.(((((((...))))))))))))).).....)))(((((..((.......))..-))))) ( -41.50) >DroPse_CAF1 62700 108 - 1 CCCUCGGCUACCCCCUGGAUCGUCCCAUCACUAACGAGCAGGAGCUGCUCCAGCUGCCGAAUGUCCACCUGCAGGAUGUGGUUAUCGUCCAGGAGAACUAAGCUUGAA------------ ...((((((...(((((((.((.((((((.(....).((((((((((...)))))...(......).)))))..)))).))....)))))))).).....))).))).------------ ( -33.00) >DroSec_CAF1 58590 120 - 1 CAUUGGGCUAUCCUCUGGACCGUCCCAUCCAUAACGAGCAGGAGCUCCUGCAGUUGACCAAUGUCCUCCUUCAGGAUGUUGUCAUCAUUCAGGAGAACUAAGUUCCCAUUAGGAAAGUUC ...(((((..(((...)))..).))))(((.......(((((....))))).(.((((.((((((((.....)))))))))))).).....)))(((((...((((.....))))))))) ( -36.90) >DroSim_CAF1 59586 119 - 1 CAUUGGGUUAUCCUCUGGACCGUCCCAUCCAUAACGAGCAGGAGCUCCUGCAGUUGACCAAUGUCCUCCUGCAGGAUGUGGUCAUCAUUCAGGAGAACUAAGUUCCCAUUAGGA-AGUUC (((((((((((....(((......)))...)))))..(((((....)))))......))))))..((((((...((((....))))...))))))....((.((((.....)))-).)). ( -38.30) >DroYak_CAF1 60074 108 - 1 CAUUGGGCUAUCCCCUGGACCGGCCCAUCCGCAACGAGCAGGAGCUCCUGCAGUUGACCAAUGUCCUGCUCCAGGAUGUGGUCAUCAUUCAGGAGAACUAAGUUCCCC------------ ...((((((.(((...)))..))))))(((((.....)).))).((((((....((((((.(((((((...))))))))))))).....)))))).............------------ ( -40.90) >DroAna_CAF1 59184 107 - 1 CACUGGGCUAUCCCCUGGACCGUCGCAUCGACAACGAGCAGGCCCUGCUGCAGCUGCCCAAUGUCCAGCUCCAGGAUGUGGUCAUCGUCCAGGAGAACUAAGCCCCA------------- ....(((((.((.(((((((....((.(((....))))).((((((.(((.(((((.........))))).))).).).))))...))))))).))....)))))..------------- ( -41.30) >consensus CAUUGGGCUAUCCCCUGGACCGUCCCAUCCAUAACGAGCAGGAGCUCCUGCAGUUGACCAAUGUCCUCCUCCAGGAUGUGGUCAUCAUUCAGGAGAACUAAGUUCCCA____________ ....(((((.((.((((((..................(((((....))))).(.((((((.((((((.....)))))))))))).).)))))).))....)))))............... (-25.14 = -25.78 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:26 2006