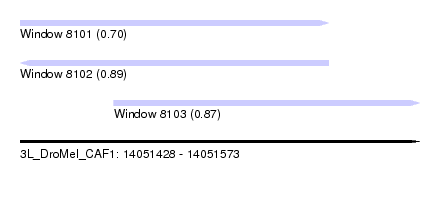
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,051,428 – 14,051,573 |
| Length | 145 |
| Max. P | 0.891968 |

| Location | 14,051,428 – 14,051,540 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 85.59 |
| Mean single sequence MFE | -24.14 |
| Consensus MFE | -16.90 |
| Energy contribution | -17.07 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14051428 112 + 23771897 AAGGAAAACGGUUUAUUAUAC------GAAUGCGAUGACAACAAAAAGGCAUAAUAGUAAAAUUAAAUGUGUCUAUUAUGCCAGGCCAAUACAAUGCAUACAUAAAACCACACUCAAA .........(((((....((.------(.(((((.((..........((((((((((....((.....))..)))))))))).........)).))))).).)))))))......... ( -19.81) >DroGri_CAF1 48457 111 + 1 -----AAACGGUUUAUUAUGCUUAUGCGAAUGUGAUGACAACAAAAAGGCAUAAUAGUAAAAUUAAAUGUGUCUAUUAUGCCAG--CAAUACAAUGCAUACAUAAAACCGCACACACA -----...((((((..((((..((((((..(((.......)))....((((((((((....((.....))..))))))))))..--........))))))))))))))))........ ( -26.90) >DroWil_CAF1 64252 102 + 1 ---CGAAACGGUUUAUUAUGC------GAAUGCGAUGACAACAAAAAGGCAUAAUAGUAAAAUUAAAUGUGUCUAUUAUGCCAG-------CAAUACACACAUAAAACCACACACAGG ---......(((((..((((.------(..(((..((....))....((((((((((....((.....))..)))))))))).)-------)).....).)))))))))......... ( -20.20) >DroMoj_CAF1 54367 111 + 1 -----AAACGGUUUAUUAUGCUUAUGCGAAUGUGAUGACAACAAAAAGGCAUAAUAGUAAAAUUAAAUGUGUCUAUUAUGCCAG--CAAUACAAUGCACACAUAAAACCACACACACA -----....(((((....(((....))).(((((.............((((((((((....((.....))..)))))))))).(--((......))).))))).)))))......... ( -27.50) >DroAna_CAF1 51413 112 + 1 UACCAGAACGGUUUAUUAUAC------GAAUACGAUGACAACAAAAAGGCAUAAUAGUAAAAUUAAAUGUGUCUAUUAUGCCGGGCCAAUACAAUGCACACAUAAAACCGCACUCACA ........((((((.((((.(------(....)))))).........((((((((((....((.....))..))))))))))......................))))))........ ( -19.90) >DroPer_CAF1 54177 117 + 1 AAUGAGAACGGUUUAUUAUACGCGUGUGAAUGCGAUGACAACAAAAAGGCAUAAUAGUAAAAUUAAAUGUGUCUAUUAUGCCA-GCCAAUACAAUGCACACAUAAAACCGCACUCGCA ...(((..((((((.((((.(((((....))))))))).........((((((((((....((.....))..)))))))))).-....................))))))..)))... ( -30.50) >consensus ____AAAACGGUUUAUUAUAC______GAAUGCGAUGACAACAAAAAGGCAUAAUAGUAAAAUUAAAUGUGUCUAUUAUGCCAG__CAAUACAAUGCACACAUAAAACCACACACACA .........(((((.................................((((((((((....((.....))..))))))))))...........(((....))).)))))......... (-16.90 = -17.07 + 0.17)
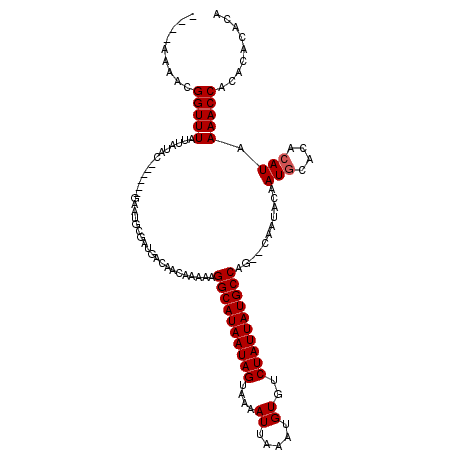

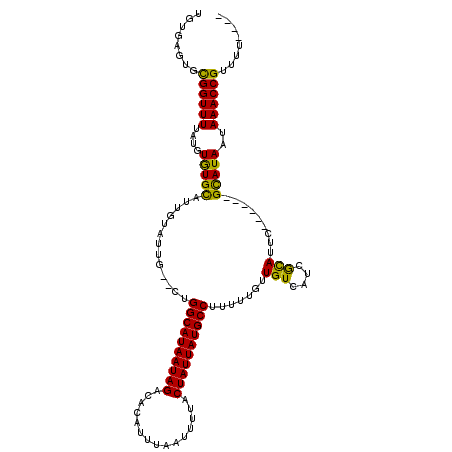
| Location | 14,051,428 – 14,051,540 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 85.59 |
| Mean single sequence MFE | -28.53 |
| Consensus MFE | -22.58 |
| Energy contribution | -21.36 |
| Covariance contribution | -1.22 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14051428 112 - 23771897 UUUGAGUGUGGUUUUAUGUAUGCAUUGUAUUGGCCUGGCAUAAUAGACACAUUUAAUUUUACUAUUAUGCCUUUUUGUUGUCAUCGCAUUC------GUAUAAUAAACCGUUUUCCUU ...(((..((((((..((((((...(((..((((..((((((((((...............))))))))))........))))..)))..)------)))))..))))))..)))... ( -27.66) >DroGri_CAF1 48457 111 - 1 UGUGUGUGCGGUUUUAUGUAUGCAUUGUAUUG--CUGGCAUAAUAGACACAUUUAAUUUUACUAUUAUGCCUUUUUGUUGUCAUCACAUUCGCAUAAGCAUAAUAAACCGUUU----- .......((((((((((((((((..(((..((--(.((((((((((...............))))))))))........).))..)))...))))..)))))..)))))))..----- ( -26.76) >DroWil_CAF1 64252 102 - 1 CCUGUGUGUGGUUUUAUGUGUGUAUUG-------CUGGCAUAAUAGACACAUUUAAUUUUACUAUUAUGCCUUUUUGUUGUCAUCGCAUUC------GCAUAAUAAACCGUUUCG--- .........(((((..((((((...((-------(.((((((((((...............))))))))))....((....))..)))..)------)))))..)))))......--- ( -25.56) >DroMoj_CAF1 54367 111 - 1 UGUGUGUGUGGUUUUAUGUGUGCAUUGUAUUG--CUGGCAUAAUAGACACAUUUAAUUUUACUAUUAUGCCUUUUUGUUGUCAUCACAUUCGCAUAAGCAUAAUAAACCGUUU----- (((.(((((((....(((((.(((......))--).((((((((((...............)))))))))).............)))))))))))).))).............----- ( -26.26) >DroAna_CAF1 51413 112 - 1 UGUGAGUGCGGUUUUAUGUGUGCAUUGUAUUGGCCCGGCAUAAUAGACACAUUUAAUUUUACUAUUAUGCCUUUUUGUUGUCAUCGUAUUC------GUAUAAUAAACCGUUCUGGUA ..(.((.(((((((....(((((...((((((((..((((((((((...............))))))))))........))))..))))..------)))))..))))))).)).).. ( -26.56) >DroPer_CAF1 54177 117 - 1 UGCGAGUGCGGUUUUAUGUGUGCAUUGUAUUGGC-UGGCAUAAUAGACACAUUUAAUUUUACUAUUAUGCCUUUUUGUUGUCAUCGCAUUCACACGCGUAUAAUAAACCGUUCUCAUU ...(((.(((((((((((((((...(((..((((-.((((((((((...............))))))))))........))))..)))....))))))))....))))))).)))... ( -38.36) >consensus UGUGAGUGCGGUUUUAUGUGUGCAUUGUAUUG__CUGGCAUAAUAGACACAUUUAAUUUUACUAUUAUGCCUUUUUGUUGUCAUCGCAUUC______GCAUAAUAAACCGUUUU____ ........((((((....(((((.............((((((((((...............)))))))))).......(((....))).........)))))..))))))........ (-22.58 = -21.36 + -1.22)

| Location | 14,051,462 – 14,051,573 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 83.06 |
| Mean single sequence MFE | -18.59 |
| Consensus MFE | -13.20 |
| Energy contribution | -13.20 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
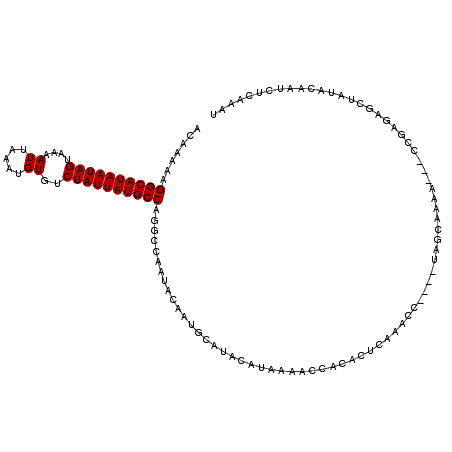
>3L_DroMel_CAF1 14051462 111 + 23771897 ACAAAAAGGCAUAAUAGUAAAAUUAAAUGUGUCUAUUAUGCCAGGCCAAUACAAUGCAUACAUAAAACCACACUCAAACC----UAGCAAAA---CCGAGAGCUAUACAAUCUCAAAU .......((((((((((....((.....))..))))))))))............(((.......................----..)))...---..((((.........)))).... ( -16.91) >DroSim_CAF1 50969 111 + 1 ACAAAAAGGCAUAAUAGUAAAAUUAAAUGUGUCUAUUAUGCCAGGCCAAUACAAUGCAUACAUAAAACCACACUCAAACC----UAGCAAAA---CCGAAAGCUAUACAAUCUCGAAU .......((((((((((....((.....))..))))))))))......................................----((((....---......))))............. ( -15.20) >DroEre_CAF1 47786 111 + 1 ACAAAAAGGCAUAAUAGUAAAAUUAAAUGUGUCUAUUAUGCCAGGCCAAUACAAUGCAUACAUAAAACCACACGCAAACC----UAGCAAAA---UUCAGAGCCAUACAAUCUCAAAU .......((((((((((....((.....))..)))))))))).((((......(((....)))..........((.....----..))....---....).))).............. ( -18.80) >DroYak_CAF1 51532 107 + 1 ACAAAAAGGCAUAAUAGUAAAAUUAAAUGUGUCUAUUAUGCCAGGCCAAUACAA----UACAUAAAACCACACUCAAACC----CAGCAAAA---CCGAGAGCUAUACAAUCUCAAAU .......((((((((((....((.....))..))))))))))............----......................----........---..((((.........)))).... ( -16.80) >DroMoj_CAF1 54402 97 + 1 ACAAAAAGGCAUAAUAGUAAAAUUAAAUGUGUCUAUUAUGCCAG--CAAUACAAUGCACACAUAAAACCACACACACACA----GAGGCU-------GAUACA--------CUCACAC .......((((((((((....((.....))..)))))))))).(--((......))).......................----(((...-------......--------))).... ( -17.90) >DroAna_CAF1 51447 118 + 1 ACAAAAAGGCAUAAUAGUAAAAUUAAAUGUGUCUAUUAUGCCGGGCCAAUACAAUGCACACAUAAAACCGCACUCACACCCACAUAGCAGGCCAGGCAGGAGCCUUGAAAUCAUAAAU .......((((((((((....((.....))..)))))))))).((((......(((....)))......((...............)).))))((((....))))............. ( -25.96) >consensus ACAAAAAGGCAUAAUAGUAAAAUUAAAUGUGUCUAUUAUGCCAGGCCAAUACAAUGCAUACAUAAAACCACACUCAAACC____UAGCAAAA___CCGAGAGCUAUACAAUCUCAAAU .......((((((((((....((.....))..))))))))))............................................................................ (-13.20 = -13.20 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:21 2006