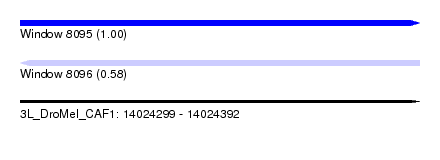
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,024,299 – 14,024,392 |
| Length | 93 |
| Max. P | 0.995033 |

| Location | 14,024,299 – 14,024,392 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 71.19 |
| Mean single sequence MFE | -26.81 |
| Consensus MFE | -9.51 |
| Energy contribution | -12.32 |
| Covariance contribution | 2.81 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.35 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.995033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
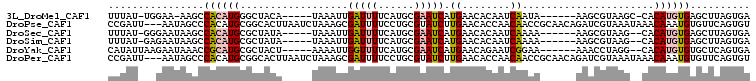
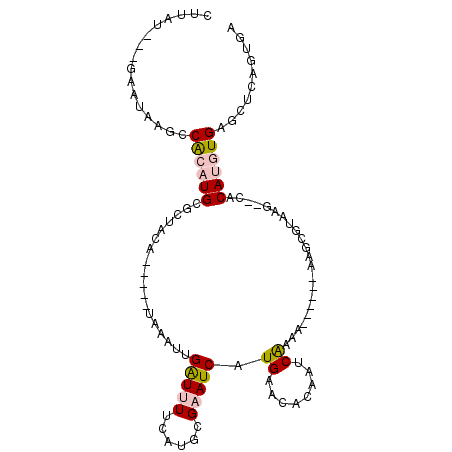
>3L_DroMel_CAF1 14024299 93 + 23771897 UCACUAAGCUCACAUGUG-GCUUACGCUU------UAUUGAUUGUGUUCAUGAUUCGCAUGAAAAUCAAUUUA-----UGUAGCCCAUGUGGCUU-UUCCA-AUAAA .....((((.((((((.(-(((.(((...------.(((((((...((((((.....)))))))))))))...-----)))))))))))))))))-.....-..... ( -29.90) >DroPse_CAF1 24101 104 + 1 ACACUGAACACAUUUGUUUAUUUACGAUCUGUUGCGGUUGUUGGUGUUCAAGAUACGCAGGAAAAUCGCUUUAGAUUAAGUGCCGCAUGUGGGCUAUU---AAUCGG .....(..(((((.((..((((((..(((((..((((((....((((.......)))).....))))))..))))))))))).)).)))))..)....---...... ( -24.40) >DroSec_CAF1 22871 93 + 1 UCACUAAGCUCACAUGUG--CUUACGCUU------UUUUGAUUGUGUUCAUGAUUCGCAUGAAAAUCAAUUUA-----UAUAGCGCAUGUGGCUUAUUCCC-AUAAA ....(((((.((((((((--((.......------..((((((...((((((.....))))))))))))....-----...))))))))))))))).....-..... ( -29.89) >DroSim_CAF1 23475 93 + 1 UCACUAAGCUCACAUGUG--CUUACGCUU------UUUUGAUUGUGUUCAUGAUUCGCAUGAAAAUCAAUUUA-----UAUAGCGCAUGUGGCUUAUUCUC-AUAAA ....(((((.((((((((--((.......------..((((((...((((((.....))))))))))))....-----...))))))))))))))).....-..... ( -29.89) >DroYak_CAF1 23583 94 + 1 UCACUGAGCACACAUGUG--CCUAGGUUU------UUCCGAUUCUGUUCAUGAUUCGCAUGAAAACCAAUUUU-----AGUAGCGCAUGCGGUUUAUUCUUAAUAUG ....(((((.(.((((((--(((((((((------((.(((.((.......)).)))...))))))).....)-----))..))))))).))))))........... ( -22.40) >DroPer_CAF1 24111 104 + 1 ACACUGAACACAUUUGUUUAUUUACGAUCUGUUGCGGUUGUUGGUGUUCAAGAUACGCAGGAAAAUCGCUUUAGAUUAAGUGCCGCAUGUGGGCUAUU---AAUCGG .....(..(((((.((..((((((..(((((..((((((....((((.......)))).....))))))..))))))))))).)).)))))..)....---...... ( -24.40) >consensus UCACUAAGCACACAUGUG__CUUACGCUU______UUUUGAUUGUGUUCAUGAUUCGCAUGAAAAUCAAUUUA_____AGUAGCGCAUGUGGCUUAUUC___AUAAA ....(((((.(((((((........(((...........(((((..((((((.....))))))...)))))..........)))))))))))))))........... ( -9.51 = -12.32 + 2.81)

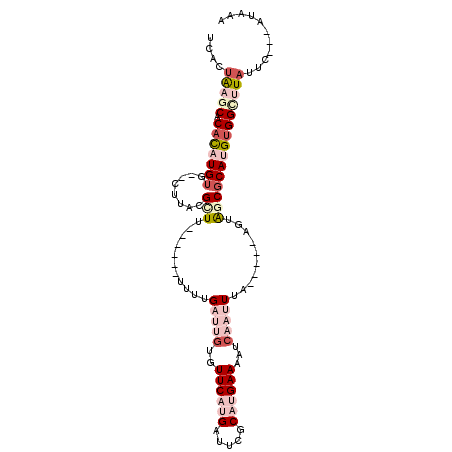
| Location | 14,024,299 – 14,024,392 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 71.19 |
| Mean single sequence MFE | -22.50 |
| Consensus MFE | -4.66 |
| Energy contribution | -5.25 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.21 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14024299 93 - 23771897 UUUAU-UGGAA-AAGCCACAUGGGCUACA-----UAAAUUGAUUUUCAUGCGAAUCAUGAACACAAUCAAUA------AAGCGUAAGC-CACAUGUGAGCUUAGUGA .....-.....-((((((((((((((((.-----...(((((((((((((.....))))))...))))))).------....)).)))-).)))))).))))..... ( -28.20) >DroPse_CAF1 24101 104 - 1 CCGAUU---AAUAGCCCACAUGCGGCACUUAAUCUAAAGCGAUUUUCCUGCGUAUCUUGAACACCAACAACCGCAACAGAUCGUAAAUAAACAAAUGUGUUCAGUGU ..((((---((..((((....).)))..))))))....(((((((...((((....(((.....)))....))))..)))))))....................... ( -19.10) >DroSec_CAF1 22871 93 - 1 UUUAU-GGGAAUAAGCCACAUGCGCUAUA-----UAAAUUGAUUUUCAUGCGAAUCAUGAACACAAUCAAAA------AAGCGUAAG--CACAUGUGAGCUUAGUGA .....-.....(((((((((((.......-----....((((((((((((.....))))))...))))))..------..((....)--).)))))).))))).... ( -24.90) >DroSim_CAF1 23475 93 - 1 UUUAU-GAGAAUAAGCCACAUGCGCUAUA-----UAAAUUGAUUUUCAUGCGAAUCAUGAACACAAUCAAAA------AAGCGUAAG--CACAUGUGAGCUUAGUGA .....-.....(((((((((((.......-----....((((((((((((.....))))))...))))))..------..((....)--).)))))).))))).... ( -24.90) >DroYak_CAF1 23583 94 - 1 CAUAUUAAGAAUAAACCGCAUGCGCUACU-----AAAAUUGGUUUUCAUGCGAAUCAUGAACAGAAUCGGAA------AAACCUAGG--CACAUGUGUGCUCAGUGA ...............(((((((....(((-----(....))))...)))))((.((.......)).))))..------...(((..(--(((....))))..)).). ( -18.80) >DroPer_CAF1 24111 104 - 1 CCGAUU---AAUAGCCCACAUGCGGCACUUAAUCUAAAGCGAUUUUCCUGCGUAUCUUGAACACCAACAACCGCAACAGAUCGUAAAUAAACAAAUGUGUUCAGUGU ..((((---((..((((....).)))..))))))....(((((((...((((....(((.....)))....))))..)))))))....................... ( -19.10) >consensus CUUAU___GAAUAAGCCACAUGCGCUACA_____UAAAUUGAUUUUCAUGCGAAUCAUGAACACAAUCAAAA______AAGCGUAAG__CACAUGUGAGCUCAGUGA ................((((((..................(((((......))))).((........))......................)))))).......... ( -4.66 = -5.25 + 0.59)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:14 2006