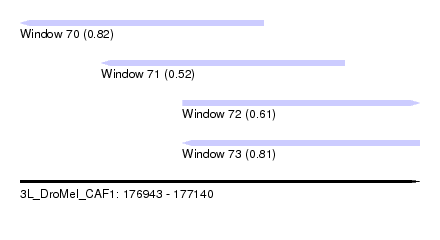
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 176,943 – 177,140 |
| Length | 197 |
| Max. P | 0.821861 |

| Location | 176,943 – 177,063 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -22.93 |
| Consensus MFE | -19.58 |
| Energy contribution | -19.80 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 176943 120 - 23771897 GACAAGCUGUUAAAUAAGAAACAGCAGCAGAUGAACCAUUCUGCUCAAGCUACACCCCCACCAAUUUCACCAACUCCACCUGCUUCUGAAACACGUAUCGUAAAAGAUCAAUGUGAUCUU .....((((((........))))))((((((........)))))).((((...............................))))......((((((((......)))..)))))..... ( -20.78) >DroSec_CAF1 10658 120 - 1 GACAAGCUGUUAAAUAAGAAACAGCAGCAGAUAAACCAUUCUGCUAAAGCUAAAUCCCCAUCAAUGUCACCAACUCCACCUGCUUCUGAAACACUUAUCGUAAAAGAUCAAUGUGAUCUU ((((.((((((........))))))((((((........))))))...................))))...................................((((((.....)))))) ( -24.90) >DroYak_CAF1 10861 120 - 1 GACAAGCUGUUGAAUAAGAAACAGCAGCAAUUGAACCAUGCUGCACAAGCUACACCACCACCAAUGUCACCAACUCCACCUGCUCCUACGACACCUAUCGUGAAAGAUCAAAGCGAUCAU ((((.(((((((.........)))))))...........(((.....)))..............))))..................(((((......)))))...((((.....)))).. ( -23.10) >consensus GACAAGCUGUUAAAUAAGAAACAGCAGCAGAUGAACCAUUCUGCUCAAGCUACACCCCCACCAAUGUCACCAACUCCACCUGCUUCUGAAACACCUAUCGUAAAAGAUCAAUGUGAUCUU ((((.((((((........)))))).(((((........)))))....................)))).....................................((((.....)))).. (-19.58 = -19.80 + 0.22)

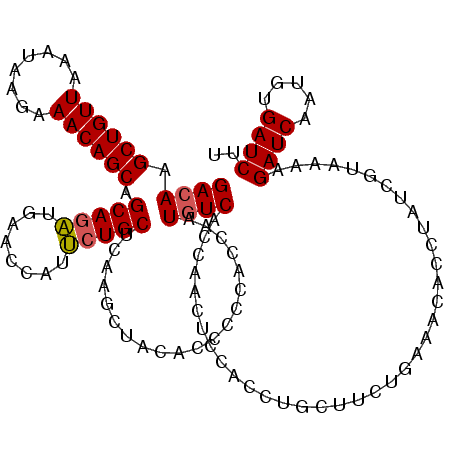
| Location | 176,983 – 177,103 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.56 |
| Mean single sequence MFE | -29.17 |
| Consensus MFE | -24.34 |
| Energy contribution | -24.90 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 176983 120 - 23771897 CCAGAGAAAAGUAGCUAGCGGAAAAGGGACUAGUUCGUUGGACAAGCUGUUAAAUAAGAAACAGCAGCAGAUGAACCAUUCUGCUCAAGCUACACCCCCACCAAUUUCACCAACUCCACC ..........(((((((((((((...((....(((.....)))..((((((........))))))..........)).)))))))..))))))........................... ( -28.50) >DroSec_CAF1 10698 120 - 1 CCAGAGAAAAGUAGCUAGCGGGAAAGCGACUAGUUCGUUGGACAAGCUGUUAAAUAAGAAACAGCAGCAGAUAAACCAUUCUGCUAAAGCUAAAUCCCCAUCAAUGUCACCAACUCCACC ..........(.((((((((......)).)))))))((((((((.((((((........))))))((((((........))))))...................)))..)))))...... ( -29.70) >DroYak_CAF1 10901 120 - 1 CCAGAGGAAGGUAGCCAGCGGAAAAGCGACUAGUUCGUUGGACAAGCUGUUGAAUAAGAAACAGCAGCAAUUGAACCAUGCUGCACAAGCUACACCACCACCAAUGUCACCAACUCCACC ...(((...(((.(((((((((..((...))..)))))))).).(((((((........))).((((((.........))))))...)))).................)))..))).... ( -29.30) >consensus CCAGAGAAAAGUAGCUAGCGGAAAAGCGACUAGUUCGUUGGACAAGCUGUUAAAUAAGAAACAGCAGCAGAUGAACCAUUCUGCUCAAGCUACACCCCCACCAAUGUCACCAACUCCACC ..........(.((((((((......)).)))))))(((((....((((((........)))))).(((((........))))).........................)))))...... (-24.34 = -24.90 + 0.56)

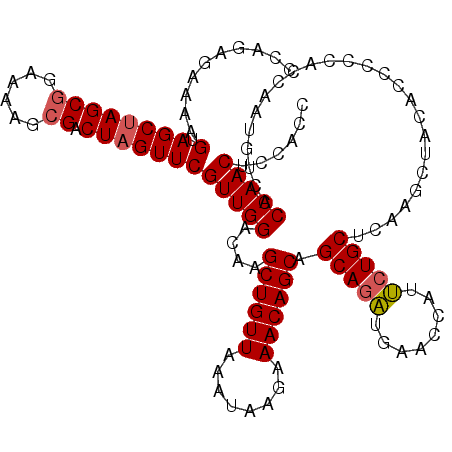
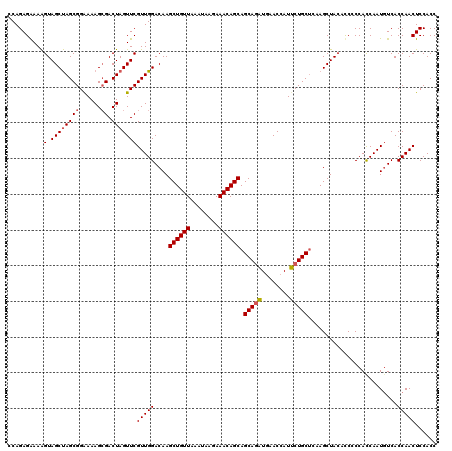
| Location | 177,023 – 177,140 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.34 |
| Mean single sequence MFE | -20.77 |
| Consensus MFE | -16.07 |
| Energy contribution | -15.63 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 177023 117 + 23771897 AAUGGUUCAUCUGCUGCUGUUUCUUAUUUAACAGCUUGUCCAACGAACUAGUCCCUUUUCCGCUAGCUACUUUUCUCUGGCUUAUUUCUUGUUCCAAAUUUGCAACCUUCUUAGGAG ..((((((....((.((((((........))))))..)).....))))))........((((((((..........))))).......((((.........))))........))). ( -20.40) >DroSec_CAF1 10738 95 + 1 AAUGGUUUAUCUGCUGCUGUUUCUUAUUUAACAGCUUGUCCAACGAACUAGUCGCUUUCCCGCUAGCUACUUUUCUCUGGCUAAUUUCUUGUUCU---------------------- ..(((.......((.((((((........))))))..)))))..((((.((..((......))((((((........))))))....)).)))).---------------------- ( -18.01) >DroYak_CAF1 10941 117 + 1 CAUGGUUCAAUUGCUGCUGUUUCUUAUUCAACAGCUUGUCCAACGAACUAGUCGCUUUUCCGCUGGCUACCUUCCUCUGGCUUAUUUCUUGUUCUAAAUUUGCCACCUUCUUAGGAG ..((((((....((.((((((........))))))..)).....))))))(((((......)).)))..(((.....((((..((((........))))..)))).......))).. ( -23.90) >consensus AAUGGUUCAUCUGCUGCUGUUUCUUAUUUAACAGCUUGUCCAACGAACUAGUCGCUUUUCCGCUAGCUACUUUUCUCUGGCUUAUUUCUUGUUCUAAAUUUGC_ACCUUCUUAGGAG ..((((((....((.((((((........))))))..)).....))))))...........(((((..........))))).................................... (-16.07 = -15.63 + -0.44)

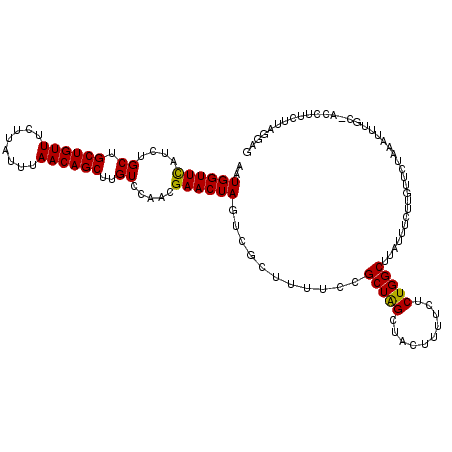
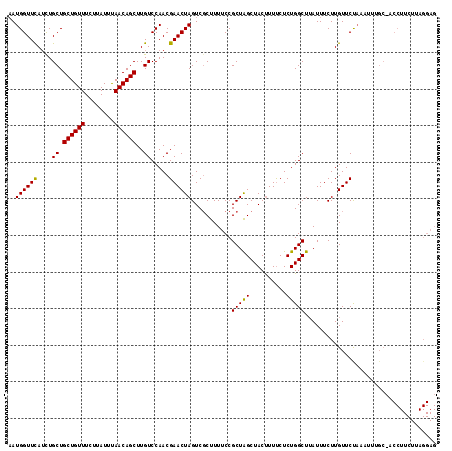
| Location | 177,023 – 177,140 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.34 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -18.24 |
| Energy contribution | -17.80 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 177023 117 - 23771897 CUCCUAAGAAGGUUGCAAAUUUGGAACAAGAAAUAAGCCAGAGAAAAGUAGCUAGCGGAAAAGGGACUAGUUCGUUGGACAAGCUGUUAAAUAAGAAACAGCAGCAGAUGAACCAUU .((((..(..((((((...(((((....(....)...))))).....))))))..).....))))....(((((((.(....((((((........))))))..).))))))).... ( -24.80) >DroSec_CAF1 10738 95 - 1 ----------------------AGAACAAGAAAUUAGCCAGAGAAAAGUAGCUAGCGGGAAAGCGACUAGUUCGUUGGACAAGCUGUUAAAUAAGAAACAGCAGCAGAUAAACCAUU ----------------------..............(((((.(((.(((.(((........))).)))..))).)))).)..((((((........))))))............... ( -17.70) >DroYak_CAF1 10941 117 - 1 CUCCUAAGAAGGUGGCAAAUUUAGAACAAGAAAUAAGCCAGAGGAAGGUAGCCAGCGGAAAAGCGACUAGUUCGUUGGACAAGCUGUUGAAUAAGAAACAGCAGCAAUUGAACCAUG .((((.......((((..((((........))))..)))).)))).(((.(((((((((..((...))..)))))))).)..(((((((.........)))))))......)))... ( -30.10) >consensus CUCCUAAGAAGGU_GCAAAUUUAGAACAAGAAAUAAGCCAGAGAAAAGUAGCUAGCGGAAAAGCGACUAGUUCGUUGGACAAGCUGUUAAAUAAGAAACAGCAGCAGAUGAACCAUU ....................................((............(((((((((..((...))..)))))))).)..((((((........)))))).))............ (-18.24 = -17.80 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:05 2006