
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,957,233 – 13,957,323 |
| Length | 90 |
| Max. P | 0.990463 |

| Location | 13,957,233 – 13,957,323 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 80.80 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -18.22 |
| Energy contribution | -20.38 |
| Covariance contribution | 2.16 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.56 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
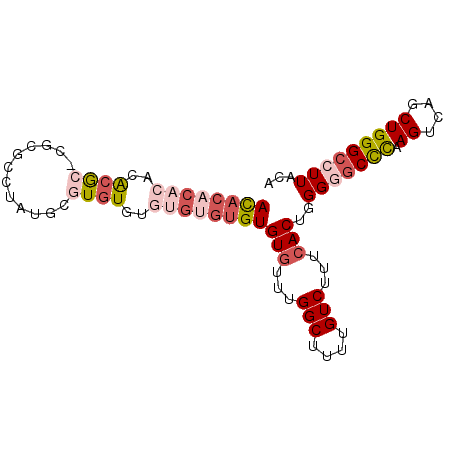
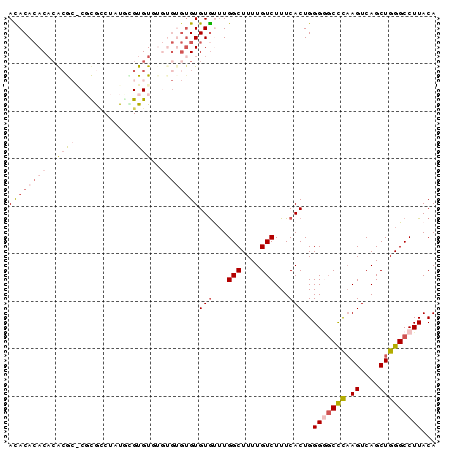
>3L_DroMel_CAF1 13957233 90 + 23771897 ACACACACACGCGCGCGCGCCUAUGAGUGUUUGUGUGUGUGUGUUUGGCUUUUGUCUUUCACUGGGGGCCCAAGUCAGCUGGGCCUUACA (((((((((((((.((((........)))).)))))))))))))..(((....)))........((((((((.(....)))))))))... ( -40.70) >DroSec_CAF1 12764 80 + 1 ACACAAACACACG--CGCGCCUAUUCGUGU--------GUGUGCAUGGCUUUUGUCUUUCACUGGGGGCCCAAGUCAGCUGGGCCUUACA .......((((((--((((......)))))--------)))))...(((....)))........((((((((.(....)))))))))... ( -29.90) >DroSim_CAF1 12856 88 + 1 ACACACACACGCG--CGCGCCUAUUUGUGUGUGUGUGUGUGUGCUUGGCUUUUGUCUUUCACUGGGAGCCCAAGUCAGCUGGGCCUUACA (((((((((((((--((((......)))))))))))))))))(((..(((((((..((((....))))..))))..)))..)))...... ( -41.20) >DroEre_CAF1 13044 88 + 1 AUACACAC--AAACACCUAGCUAUGCGUGUGUGUGUGUGUGUAUUUGGCUUUUGUCUUUCACUGGGGGCUUCAGUCAGCUGGGCCUUACA ((((((((--(.((((...((...))..)))).)))))))))....(((....)))........((((((.(((....)))))))))... ( -28.90) >DroYak_CAF1 13269 88 + 1 AUACACACACACACACCUAUCUAUAUGUGUUC--GCGUGUGUGUUUGGCUUUUGUCUUUCACUGGGCUCCCAAGUCAGCUGGGCCUUACA ..((((((((.(((((.((....)).))))..--).))))))))..((((((((.((.....(((....))))).)))..)))))..... ( -23.30) >consensus ACACACACACACGC_CGCGCCUAUGCGUGUGUGUGUGUGUGUGUUUGGCUUUUGUCUUUCACUGGGGGCCCAAGUCAGCUGGGCCUUACA ((((((((..((((............))))..))))))))(((...(((....)))...)))..(((((((.((....)))))))))... (-18.22 = -20.38 + 2.16)

| Location | 13,957,233 – 13,957,323 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 80.80 |
| Mean single sequence MFE | -26.18 |
| Consensus MFE | -16.68 |
| Energy contribution | -17.44 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
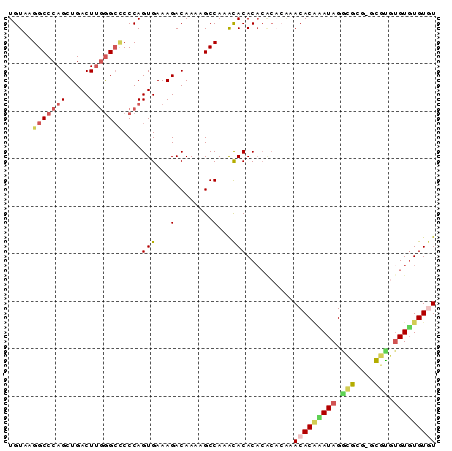
>3L_DroMel_CAF1 13957233 90 - 23771897 UGUAAGGCCCAGCUGACUUGGGCCCCCAGUGAAAGACAAAAGCCAAACACACACACACAAACACUCAUAGGCGCGCGCGCGUGUGUGUGU .....(((((((.....)))))))........................((((((((((.....(.....)(((....))))))))))))) ( -30.40) >DroSec_CAF1 12764 80 - 1 UGUAAGGCCCAGCUGACUUGGGCCCCCAGUGAAAGACAAAAGCCAUGCACAC--------ACACGAAUAGGCGCG--CGUGUGUUUGUGU .....(((((((.....)))))))....(((...(........)...)))..--------(((((((((.((...--.)).))))))))) ( -24.40) >DroSim_CAF1 12856 88 - 1 UGUAAGGCCCAGCUGACUUGGGCUCCCAGUGAAAGACAAAAGCCAAGCACACACACACACACACAAAUAGGCGCG--CGCGUGUGUGUGU .....(((((((.....)))))))......................((((((((((.(.(.(.(......).).)--.).)))))))))) ( -26.40) >DroEre_CAF1 13044 88 - 1 UGUAAGGCCCAGCUGACUGAAGCCCCCAGUGAAAGACAAAAGCCAAAUACACACACACACACACGCAUAGCUAGGUGUUU--GUGUGUAU .....(((....((.((((.......))))...))......)))..(((((((((.((((....((...))...)))).)--)))))))) ( -23.30) >DroYak_CAF1 13269 88 - 1 UGUAAGGCCCAGCUGACUUGGGAGCCCAGUGAAAGACAAAAGCCAAACACACACGC--GAACACAUAUAGAUAGGUGUGUGUGUGUGUAU .....(((....((..(((((....)))).)..))......)))..((((((((((--..((((..........)))))))))))))).. ( -26.40) >consensus UGUAAGGCCCAGCUGACUUGGGCCCCCAGUGAAAGACAAAAGCCAAACACACACACACAAACACAAAUAGGCGCG_GCGUGUGUGUGUGU .....(((((((.....)))))))....(((...(........)...)))..........(((((((((.(((....))).))))))))) (-16.68 = -17.44 + 0.76)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:41 2006