
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,947,225 – 13,947,422 |
| Length | 197 |
| Max. P | 0.999595 |

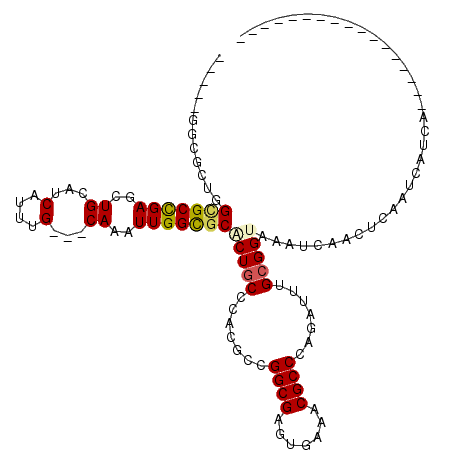
| Location | 13,947,225 – 13,947,325 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.40 |
| Mean single sequence MFE | -34.57 |
| Consensus MFE | -18.45 |
| Energy contribution | -18.59 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.546247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13947225 100 - 23771897 GAUUCGCCACUGGCGCCGAGCUGCAUCACCUG---CAAAUUGGCGCACUGCCCACGCCGGCGAGUGAAACGCCACGAUUUGCGGUAAAUCAACUCAAGCAUCA----------------- ((((..((....(((((((..((((.....))---))..)))))))...((...((..((((.......)))).))....))))..)))).............----------------- ( -33.50) >DroVir_CAF1 2761 97 - 1 -----GGUGUCGGUGCCGAGCUGCAUCAUUUG---CAAAUUGGCGCCCUGCCGACGCCGGCGAGUGAAACGCCAAGAUUUGCGGUAAAUCAACUC----AUCA------GUCAGC----- -----((((((((((((((..((((.....))---))..))))).....)))))))))(((..((((..(((........)))((......))))----))..------)))...----- ( -36.70) >DroPse_CAF1 2728 89 - 1 -----------GGCGCCGAGCUGCAUCACUUG---CAAAUUGGCGCACUCCCCACGCCGGCGAGUGAAACGCCCCGAUUUGCGGUAAAUCAACUCAAUCAUCA----------------- -----------((((((((..((((.....))---))..))))).(((((.((.....)).)))))....)))(((.....)))...................----------------- ( -29.10) >DroGri_CAF1 2436 99 - 1 -----GGCGGUGGUGCCGAGAUGCAUCAAUUG---CAAAUUGGCGCCCUGCCCACGCCGGCGAGUGAAACGCCCAGAUUUGCGGUAAAUCAACACAGUCAUCA------GUCA------- -----(((((.((((((((..((((.....))---))..)))))))))))))......(((..((((..(((........)))((......))....))))..------))).------- ( -37.70) >DroWil_CAF1 3738 98 - 1 -----GGCGCUGGAGCUGAGCUGCAUCAUUUGUUACAAAUUGGUGCGCUGCCCACGCCGGCGAGUGAAACGCCCAGAUUUGCGGUAAAUCAAACCAAUCAUCC----------------- -----((((.(((.((..(((.((((((.((......)).))))))))))))))))))((((.......))))..((((...(((.......)))))))....----------------- ( -32.60) >DroAna_CAF1 3355 117 - 1 GCUAUGCCACUGGCGCCGAGCUGCAGCAUCUG---CAAAUUGGCGCACUGCCCACGCCGGCGAGUGAAACGCCCCGAUUUGCGGUAAAUCAACUCAAUCAUCCAACUCAGUCGCCGAGAC ((...((.....(((((((..(((((...)))---))..)))))))...))....))((((((.(((..(((........)))((......)).............))).)))))).... ( -37.80) >consensus _____GGCGCUGGCGCCGAGCUGCAUCAUUUG___CAAAUUGGCGCACUGCCCACGCCGGCGAGUGAAACGCCCAGAUUUGCGGUAAAUCAACUCAAUCAUCA_________________ ............(((((((..((...(....)...))..)))))))(((((.......((((.......)))).......)))))................................... (-18.45 = -18.59 + 0.14)


| Location | 13,947,248 – 13,947,362 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 96.23 |
| Mean single sequence MFE | -55.18 |
| Consensus MFE | -49.99 |
| Energy contribution | -50.02 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13947248 114 + 23771897 AAAUCGUGGCGUUUCACUCGCCGGCGUGGGCAGUGCGCCAAUUUGCAGGUGAUGCAGCUCGGCGCCAGUGGCGAAUCCGCCGCUGGAGUCGCCGCCAUUGGUGUCGGCAUCG---CC ......(((((...((((.(((......)))))))))))).......((((((((....(((((((((((((((.((((....)))).)))))))...))))))))))))))---)) ( -56.30) >DroSec_CAF1 2913 114 + 1 AAAUCGUGGCGUUUCACUCGCCGGCGUGGGCAGUGCGCCAAUUUGCAGGUGAUGCAACUCGGCGCCAGUGGCGAAUCCGCCGCUGGAGUCGCCGCCAUUGGUGUCGGCAUCG---CC ......(((((...((((.(((......)))))))))))).......((((((((....(((((((((((((((.((((....)))).)))))))...))))))))))))))---)) ( -56.30) >DroSim_CAF1 2942 114 + 1 AAAUCGUGGCGUUUCACUCGCCGGCGUGGGCAGUGCGCCAAUUUGCAGGUGAUGCAGCUCGGCGCCAGUGGCGAAUCCGCCGCUGGAGUCGCCGCCAUUGGUGUCGGCAUCG---CC ......(((((...((((.(((......)))))))))))).......((((((((....(((((((((((((((.((((....)))).)))))))...))))))))))))))---)) ( -56.30) >DroEre_CAF1 2874 114 + 1 AAAUCGUGGCGUUUCACUCGCCGGCGUGGGCAGUGCGCCAAUUUGCAGGUGAUGCAGCUCGGCGCCAGUGGCGAAUCCGCCGCUGGAGUCGCCGCCAUUGGUGUCGGCAUCG---CC ......(((((...((((.(((......)))))))))))).......((((((((....(((((((((((((((.((((....)))).)))))))...))))))))))))))---)) ( -56.30) >DroYak_CAF1 2906 117 + 1 AAAUCGUGGCGUUUCACUCGCCGGCGUGGGCAGUGCGCCAAUUUGCAGGUGAUGCAGCUCGGCGCCAGUGGCGAAUCCGCCGCUGGAGUCGCCGCCAUUGGUGUCGGCAUCGUAACC .(((.((((((((.((.(((((.(((..(((.....)))....))).))))))).)))..(((.(((((((((....))))))))).)))))))).)))((((....))))...... ( -52.80) >DroAna_CAF1 3395 114 + 1 AAAUCGGGGCGUUUCACUCGCCGGCGUGGGCAGUGCGCCAAUUUGCAGAUGCUGCAGCUCGGCGCCAGUGGCAUAGCAGCCGCUGGAGUCGCCGCCAAUGGUGUCAGCAUCG---CC .......((((...((((.(((......))))))))))).....((.(((((((((.(.(((((((((((((......))))))))...))))).....).)).))))))))---). ( -53.10) >consensus AAAUCGUGGCGUUUCACUCGCCGGCGUGGGCAGUGCGCCAAUUUGCAGGUGAUGCAGCUCGGCGCCAGUGGCGAAUCCGCCGCUGGAGUCGCCGCCAUUGGUGUCGGCAUCG___CC .......((((.......))))((((..(((...(((((...(((((.....)))))...)))))((((((((....))))))))..)))..))))...((((....))))...... (-49.99 = -50.02 + 0.03)


| Location | 13,947,248 – 13,947,362 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 96.23 |
| Mean single sequence MFE | -50.75 |
| Consensus MFE | -45.82 |
| Energy contribution | -45.85 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13947248 114 - 23771897 GG---CGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCGGAUUCGCCACUGGCGCCGAGCUGCAUCACCUGCAAAUUGGCGCACUGCCCACGCCGGCGAGUGAAACGCCACGAUUU ((---((..((((.......))))((((....(((.((((....)))).)))(((((((..((((.....))))..)))))))..))))..))))((((.......))))....... ( -51.50) >DroSec_CAF1 2913 114 - 1 GG---CGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCGGAUUCGCCACUGGCGCCGAGUUGCAUCACCUGCAAAUUGGCGCACUGCCCACGCCGGCGAGUGAAACGCCACGAUUU ((---((..((((.......))))((((....(((.((((....)))).)))(((((((.(((((.....))))).)))))))..))))..))))((((.......))))....... ( -52.10) >DroSim_CAF1 2942 114 - 1 GG---CGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCGGAUUCGCCACUGGCGCCGAGCUGCAUCACCUGCAAAUUGGCGCACUGCCCACGCCGGCGAGUGAAACGCCACGAUUU ((---((..((((.......))))((((....(((.((((....)))).)))(((((((..((((.....))))..)))))))..))))..))))((((.......))))....... ( -51.50) >DroEre_CAF1 2874 114 - 1 GG---CGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCGGAUUCGCCACUGGCGCCGAGCUGCAUCACCUGCAAAUUGGCGCACUGCCCACGCCGGCGAGUGAAACGCCACGAUUU ((---((..((((.......))))((((....(((.((((....)))).)))(((((((..((((.....))))..)))))))..))))..))))((((.......))))....... ( -51.50) >DroYak_CAF1 2906 117 - 1 GGUUACGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCGGAUUCGCCACUGGCGCCGAGCUGCAUCACCUGCAAAUUGGCGCACUGCCCACGCCGGCGAGUGAAACGCCACGAUUU (((......)))........(((((.(.(((((((.((((....)))).)))(((((((..((((.....))))..)))))))...(((......))))))))...)))))...... ( -47.60) >DroAna_CAF1 3395 114 - 1 GG---CGAUGCUGACACCAUUGGCGGCGACUCCAGCGGCUGCUAUGCCACUGGCGCCGAGCUGCAGCAUCUGCAAAUUGGCGCACUGCCCACGCCGGCGAGUGAAACGCCCCGAUUU ((---((..((..(.....)..))((((....(((.(((......))).)))(((((((..(((((...)))))..)))))))..))))..))))((((.......))))....... ( -50.30) >consensus GG___CGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCGGAUUCGCCACUGGCGCCGAGCUGCAUCACCUGCAAAUUGGCGCACUGCCCACGCCGGCGAGUGAAACGCCACGAUUU .....((..((((.......))))((((....(((.((((....)))).)))(((((((..((((.....))))..)))))))........))))((((.......)))).)).... (-45.82 = -45.85 + 0.03)
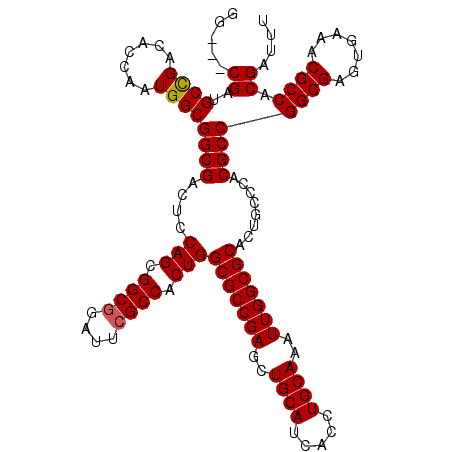

| Location | 13,947,288 – 13,947,390 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 98.06 |
| Mean single sequence MFE | -44.76 |
| Consensus MFE | -42.68 |
| Energy contribution | -42.68 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.59 |
| SVM RNA-class probability | 0.999420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13947288 102 + 23771897 AUUUGCAGGUGAUGCAGCUCGGCGCCAGUGGCGAAUCCGCCGCUGGAGUCGCCGCCAUUGGUGUCGGCAUCG---CCGUAACCACCAACCACACCGACAUGGCCA .......(((.((((....((((((((((((((....)))))))))...)))))...((((((.((((...)---)))....)))))).......).))).))). ( -45.20) >DroSec_CAF1 2953 102 + 1 AUUUGCAGGUGAUGCAACUCGGCGCCAGUGGCGAAUCCGCCGCUGGAGUCGCCGCCAUUGGUGUCGGCAUCG---CCGUAACCACCAACCACACCGACAUGGCCA .......(((.((((....((((((((((((((....)))))))))...)))))...((((((.((((...)---)))....)))))).......).))).))). ( -45.20) >DroSim_CAF1 2982 102 + 1 AUUUGCAGGUGAUGCAGCUCGGCGCCAGUGGCGAAUCCGCCGCUGGAGUCGCCGCCAUUGGUGUCGGCAUCG---CCGUAACCACCAACCACACCGACAUGGCCA .......(((.((((....((((((((((((((....)))))))))...)))))...((((((.((((...)---)))....)))))).......).))).))). ( -45.20) >DroEre_CAF1 2914 102 + 1 AUUUGCAGGUGAUGCAGCUCGGCGCCAGUGGCGAAUCCGCCGCUGGAGUCGCCGCCAUUGGUGUCGGCAUCG---CCGUAACCACCAACCACACCGACAUGGCCA .......(((.((((....((((((((((((((....)))))))))...)))))...((((((.((((...)---)))....)))))).......).))).))). ( -45.20) >DroYak_CAF1 2946 105 + 1 AUUUGCAGGUGAUGCAGCUCGGCGCCAGUGGCGAAUCCGCCGCUGGAGUCGCCGCCAUUGGUGUCGGCAUCGUAACCGUAACCACCAACCACACCGACAUGGCCG .......(((.((((....((((((((((((((....)))))))))...)))))...((((((.(((........)))....)))))).......).))).))). ( -43.00) >consensus AUUUGCAGGUGAUGCAGCUCGGCGCCAGUGGCGAAUCCGCCGCUGGAGUCGCCGCCAUUGGUGUCGGCAUCG___CCGUAACCACCAACCACACCGACAUGGCCA .......(((.((((....((((((((((((((....)))))))))...)))))...((((((.(((........)))....)))))).......).))).))). (-42.68 = -42.68 + -0.00)



| Location | 13,947,288 – 13,947,390 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 98.06 |
| Mean single sequence MFE | -48.00 |
| Consensus MFE | -45.08 |
| Energy contribution | -44.92 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.997590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13947288 102 - 23771897 UGGCCAUGUCGGUGUGGUUGGUGGUUACGG---CGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCGGAUUCGCCACUGGCGCCGAGCUGCAUCACCUGCAAAU .(((...((((.(((.(((((((....(((---(...)))).))))))).))).)))).((((.((((....)))).)))).)))....((((.....))))... ( -47.40) >DroSec_CAF1 2953 102 - 1 UGGCCAUGUCGGUGUGGUUGGUGGUUACGG---CGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCGGAUUCGCCACUGGCGCCGAGUUGCAUCACCUGCAAAU .(((...((((.(((.(((((((....(((---(...)))).))))))).))).)))).((((.((((....)))).)))).)))...(((((.....))))).. ( -48.40) >DroSim_CAF1 2982 102 - 1 UGGCCAUGUCGGUGUGGUUGGUGGUUACGG---CGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCGGAUUCGCCACUGGCGCCGAGCUGCAUCACCUGCAAAU .(((...((((.(((.(((((((....(((---(...)))).))))))).))).)))).((((.((((....)))).)))).)))....((((.....))))... ( -47.40) >DroEre_CAF1 2914 102 - 1 UGGCCAUGUCGGUGUGGUUGGUGGUUACGG---CGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCGGAUUCGCCACUGGCGCCGAGCUGCAUCACCUGCAAAU .(((...((((.(((.(((((((....(((---(...)))).))))))).))).)))).((((.((((....)))).)))).)))....((((.....))))... ( -47.40) >DroYak_CAF1 2946 105 - 1 CGGCCAUGUCGGUGUGGUUGGUGGUUACGGUUACGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCGGAUUCGCCACUGGCGCCGAGCUGCAUCACCUGCAAAU ((((...((((.(((.(((((((....((((......)))).))))))).))).)))).((((.((((....)))).)))).))))...((((.....))))... ( -49.40) >consensus UGGCCAUGUCGGUGUGGUUGGUGGUUACGG___CGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCGGAUUCGCCACUGGCGCCGAGCUGCAUCACCUGCAAAU ((((...((((.(((.(((((((....(((........))).))))))).))).)))).((((.((((....)))).)))).))))...((((.....))))... (-45.08 = -44.92 + -0.16)

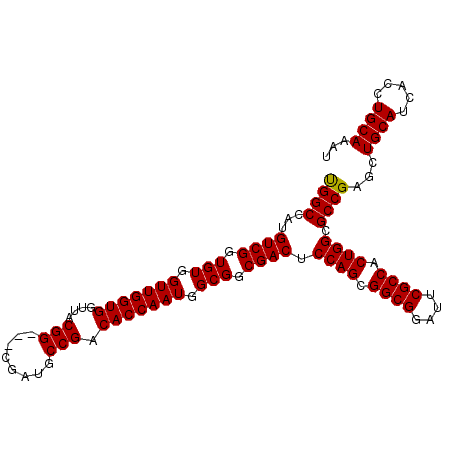

| Location | 13,947,325 – 13,947,422 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 94.26 |
| Mean single sequence MFE | -49.86 |
| Consensus MFE | -42.30 |
| Energy contribution | -43.10 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.63 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.76 |
| SVM RNA-class probability | 0.999595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13947325 97 - 23771897 GGUGUUGGUGGCUACGGCUACAGCUACGGCUAUGGCCAUGUCGGUGUGGUUGGUGGUUACGG---CGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCG (.((((((.......(((((.(((....))).)))))..((((.(((.(((((((....(((---(...)))).))))))).))).)))).)))))).). ( -49.70) >DroSec_CAF1 2990 97 - 1 GGUGUUGGUGGCUACGGCUACAGCUACGGCUAUGGCCAUGUCGGUGUGGUUGGUGGUUACGG---CGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCG (.((((((.......(((((.(((....))).)))))..((((.(((.(((((((....(((---(...)))).))))))).))).)))).)))))).). ( -49.70) >DroSim_CAF1 3019 97 - 1 GGUGUUGGUGGCUACGGCUACAGCUACGGCUAUGGCCAUGUCGGUGUGGUUGGUGGUUACGG---CGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCG (.((((((.......(((((.(((....))).)))))..((((.(((.(((((((....(((---(...)))).))))))).))).)))).)))))).). ( -49.70) >DroEre_CAF1 2951 91 - 1 GGUGUUGGUGGCUACGGC------UACGGCUAUGGCCAUGUCGGUGUGGUUGGUGGUUACGG---CGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCG (.((((((((((((..((------....))..)))))).((((.(((.(((((((....(((---(...)))).))))))).))).)))).)))))).). ( -50.60) >DroYak_CAF1 2983 94 - 1 GGUGUUGGUAGCUACGGC------UACGGCUACGGCCAUGUCGGUGUGGUUGGUGGUUACGGUUACGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCG (.((((((((((....))------)))(((....)))..((((.(((.(((((((....((((......)))).))))))).))).))))..))))).). ( -49.60) >consensus GGUGUUGGUGGCUACGGCUACAGCUACGGCUAUGGCCAUGUCGGUGUGGUUGGUGGUUACGG___CGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCG (.((((((((((((.((((........)))).)))))).((((.(((.(((((((....(((........))).))))))).))).)))).)))))).). (-42.30 = -43.10 + 0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:33 2006