| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,904,782 – 13,904,907 |
| Length | 125 |
| Max. P | 0.988271 |

| Location | 13,904,782 – 13,904,877 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 78.22 |
| Mean single sequence MFE | -27.83 |
| Consensus MFE | -18.11 |
| Energy contribution | -18.50 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13904782 95 + 23771897 ---GAUAUAU--AG-GGCCAGACUGUAGCGCGCUUGCGAAAGUGGAAAGUAUCUGCUGGAUACUUUGGAUACUUUCCGUUUUGGGGGAAUUAAUUAGACCA ---(...(((--((-.......)))))...).(((.(((((.(((((((((((((..(....)..)))))))))))))))))).))).............. ( -26.20) >DroPse_CAF1 13565 92 + 1 ---GAAAAAU--AA-ACCCAAACUGAAGUUCAUUUGCGAAAGUGGAAAGUAUCCG---UAUCCGUCGGAUACUUUCCGUUUUUGGAGAAUUAACUAGACUA ---.......--..-...........((((.((((.(((((((((((((((((((---.......)))))))))))))))))))..)))).))))...... ( -28.80) >DroSec_CAF1 13069 95 + 1 ---GAUAUAU--AG-GGCCAGACUGUAGCGCGCUUGCGAAAGUGGAAAGUAUCUGCUGGAUACUUUAGAUACUUUCCGUUUUGGGGGAAUUAAUUAGACCA ---(...(((--((-.......)))))...).(((.(((((.(((((((((((((..(....)..)))))))))))))))))).))).............. ( -27.50) >DroYak_CAF1 14252 97 + 1 AAAAAAAUAU--AG-GGCCAGACUGUAGCGCGCUUGCGAAAGUGGAAAGUAUCUGCUGGAUACUUUGGAUACUUUCCGUU-UGGGGGAAUUAAUUAGACCA ..........--..-..((((((.....(((....))).....((((((((((((..(....)..)))))))))))))))-)))((............)). ( -27.20) >DroAna_CAF1 14885 78 + 1 ---GAAAAAUCCAAAACCCAA----------ACUUGCGAAAGUGGAAAGUAUCUGAUGGAU---------ACUUUCCGUU-UGGGGGAAUUAAUUAGACCA ---......(((....(((((----------((..((....))((((((((((.....)))---------))))))))))-)))))))............. ( -29.30) >DroPer_CAF1 13549 92 + 1 ---GAAAAAU--AA-ACCCAAACUGAAGUUCGCUUGCGAAAGUGGAAAGUAUCCG---UAUCCGUCGGAUACUUUCCGUUUUUGGAGAAUUAAUUAGACUA ---.......--..-.......(((((((((.....(((((((((((((((((((---.......)))))))))))))))))))..)))))..)))).... ( -28.00) >consensus ___GAAAAAU__AA_ACCCAAACUGUAGCGCGCUUGCGAAAGUGGAAAGUAUCUGCUGGAUACUUUGGAUACUUUCCGUUUUGGGGGAAUUAAUUAGACCA .................((................((....))((((((((((((..........))))))))))))......))................ (-18.11 = -18.50 + 0.39)



| Location | 13,904,782 – 13,904,877 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 78.22 |
| Mean single sequence MFE | -22.93 |
| Consensus MFE | -12.02 |
| Energy contribution | -12.52 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13904782 95 - 23771897 UGGUCUAAUUAAUUCCCCCAAAACGGAAAGUAUCCAAAGUAUCCAGCAGAUACUUUCCACUUUCGCAAGCGCGCUACAGUCUGGCC-CU--AUAUAUC--- .((((..(((..............((((((((((..............)))))))))).....(((....)))....)))..))))-..--.......--- ( -20.24) >DroPse_CAF1 13565 92 - 1 UAGUCUAGUUAAUUCUCCAAAAACGGAAAGUAUCCGACGGAUA---CGGAUACUUUCCACUUUCGCAAAUGAACUUCAGUUUGGGU-UU--AUUUUUC--- ........................((((((((((((.......---))))))))))))........((((((((((......))))-))--))))...--- ( -24.90) >DroSec_CAF1 13069 95 - 1 UGGUCUAAUUAAUUCCCCCAAAACGGAAAGUAUCUAAAGUAUCCAGCAGAUACUUUCCACUUUCGCAAGCGCGCUACAGUCUGGCC-CU--AUAUAUC--- .((((..(((..............(((((((((((...((.....))))))))))))).....(((....)))....)))..))))-..--.......--- ( -21.90) >DroYak_CAF1 14252 97 - 1 UGGUCUAAUUAAUUCCCCCA-AACGGAAAGUAUCCAAAGUAUCCAGCAGAUACUUUCCACUUUCGCAAGCGCGCUACAGUCUGGCC-CU--AUAUUUUUUU .((((..(((..........-...((((((((((..............)))))))))).....(((....)))....)))..))))-..--.......... ( -20.24) >DroAna_CAF1 14885 78 - 1 UGGUCUAAUUAAUUCCCCCA-AACGGAAAGU---------AUCCAUCAGAUACUUUCCACUUUCGCAAGU----------UUGGGUUUUGGAUUUUUC--- .(((((((........((((-((((((((((---------(((.....)))))))))).(....)...))----------)))))..)))))))....--- ( -23.30) >DroPer_CAF1 13549 92 - 1 UAGUCUAAUUAAUUCUCCAAAAACGGAAAGUAUCCGACGGAUA---CGGAUACUUUCCACUUUCGCAAGCGAACUUCAGUUUGGGU-UU--AUUUUUC--- ...............((((((...((((((((((((.......---))))))))))))...((((....))))......)))))).-..--.......--- ( -27.00) >consensus UGGUCUAAUUAAUUCCCCCAAAACGGAAAGUAUCCAAAGUAUCCAGCAGAUACUUUCCACUUUCGCAAGCGCGCUACAGUCUGGCC_CU__AUAUUUC___ ........................((((((((((..............))))))))))........................................... (-12.02 = -12.52 + 0.50)

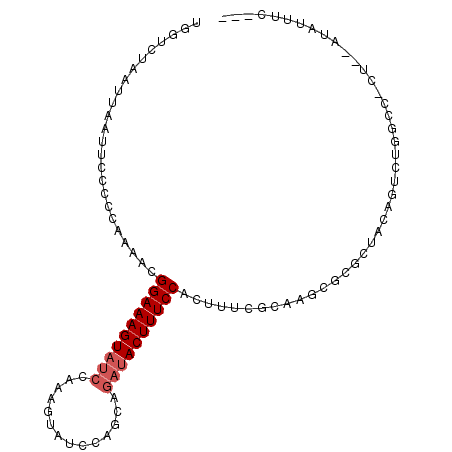
| Location | 13,904,810 – 13,904,907 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 83.48 |
| Mean single sequence MFE | -26.41 |
| Consensus MFE | -19.12 |
| Energy contribution | -20.07 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
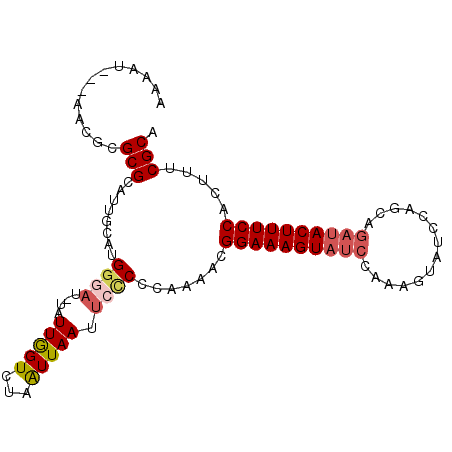
>3L_DroMel_CAF1 13904810 97 + 23771897 UGCGAAAGUGGAAAGUAUCUGCUGGAUACUUUGGAUACUUUCCGUUUUGGGGGAAUUAAUUAGACCAAUAUAUCCCAUGCAAUCCGCGCGUU---AUUUU .(((.....((((((((((((..(....)..))))))))))))(..(((.((((..................))))...)))..).)))...---..... ( -25.97) >DroPse_CAF1 13593 94 + 1 UGCGAAAGUGGAAAGUAUCCG---UAUCCGUCGGAUACUUUCCGUUUUUGGAGAAUUAACUAGACUAA---AUCCCAUGCAAAGCGCGCGUUUAUAUUUU .(((...((((((((((((((---.......))))))))))))((...(((.((.(((.......)))---.))))).))...)).)))........... ( -29.20) >DroSec_CAF1 13097 97 + 1 UGCGAAAGUGGAAAGUAUCUGCUGGAUACUUUAGAUACUUUCCGUUUUGGGGGAAUUAAUUAGACCAAUAUAUCCCAUGCAAUCCGCGCGUU---AUUUU .(((.....((((((((((((..(....)..))))))))))))(..(((.((((..................))))...)))..).)))...---..... ( -27.27) >DroEre_CAF1 13347 96 + 1 UGCGAAAGUGGAAAGUAUCUGCUGGAUACUUUGGAUACUUUCCGUUUUGGGGGAAUUAAUUAGACCAAUAUAUG-CAUGCAAUCCGCGCGUU---AUUUU .((((((((((((((((((((..(....)..)))))))))))))))))((((............))......((-....))..)).)))...---..... ( -23.60) >DroAna_CAF1 14906 88 + 1 UGCGAAAGUGGAAAGUAUCUGAUGGAU---------ACUUUCCGUU-UGGGGGAAUUAAUUAGACCAAAA-A-UCCAUGCAAAGCGCGCGUUUAUUUUUU .(((...((((((((((((.....)))---------)))))))((.-((((...................-.-)))).))...)).)))........... ( -23.25) >DroPer_CAF1 13577 93 + 1 UGCGAAAGUGGAAAGUAUCCG---UAUCCGUCGGAUACUUUCCGUUUUUGGAGAAUUAAUUAGACUAA---AUCCCAUGCAAAGCGCGCGUUUAU-UUUU .(((...((((((((((((((---.......))))))))))))((...(((.((.(((.......)))---.))))).))...)).)))......-.... ( -29.20) >consensus UGCGAAAGUGGAAAGUAUCUGCUGGAUACUUUGGAUACUUUCCGUUUUGGGGGAAUUAAUUAGACCAAUA_AUCCCAUGCAAACCGCGCGUU___AUUUU .(((.....((((((((((((..........))))))))))))((..(((((....................))))).))....)))............. (-19.12 = -20.07 + 0.95)


| Location | 13,904,810 – 13,904,907 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 83.48 |
| Mean single sequence MFE | -24.35 |
| Consensus MFE | -15.98 |
| Energy contribution | -16.62 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13904810 97 - 23771897 AAAAU---AACGCGCGGAUUGCAUGGGAUAUAUUGGUCUAAUUAAUUCCCCCAAAACGGAAAGUAUCCAAAGUAUCCAGCAGAUACUUUCCACUUUCGCA .....---.....(((((.....((((....((((((...))))))...))))....((((((((((..............))))))))))...))))). ( -24.34) >DroPse_CAF1 13593 94 - 1 AAAAUAUAAACGCGCGCUUUGCAUGGGAU---UUAGUCUAGUUAAUUCUCCAAAAACGGAAAGUAUCCGACGGAUA---CGGAUACUUUCCACUUUCGCA .............(((.......(((((.---((((.....)))).)).))).....((((((((((((.......---)))))))))))).....))). ( -27.50) >DroSec_CAF1 13097 97 - 1 AAAAU---AACGCGCGGAUUGCAUGGGAUAUAUUGGUCUAAUUAAUUCCCCCAAAACGGAAAGUAUCUAAAGUAUCCAGCAGAUACUUUCCACUUUCGCA .....---.....(((((.....((((....((((((...))))))...))))....(((((((((((...((.....)))))))))))))...))))). ( -26.00) >DroEre_CAF1 13347 96 - 1 AAAAU---AACGCGCGGAUUGCAUG-CAUAUAUUGGUCUAAUUAAUUCCCCCAAAACGGAAAGUAUCCAAAGUAUCCAGCAGAUACUUUCCACUUUCGCA .....---.....(((((.......-......((((..............))))...((((((((((..............))))))))))...))))). ( -20.08) >DroAna_CAF1 14906 88 - 1 AAAAAAUAAACGCGCGCUUUGCAUGGA-U-UUUUGGUCUAAUUAAUUCCCCCA-AACGGAAAGU---------AUCCAUCAGAUACUUUCCACUUUCGCA ...........(((.(.((((...(((-.-..(((((...))))).)))..))-)))(((((((---------(((.....)))))))))).....))). ( -20.30) >DroPer_CAF1 13577 93 - 1 AAAA-AUAAACGCGCGCUUUGCAUGGGAU---UUAGUCUAAUUAAUUCUCCAAAAACGGAAAGUAUCCGACGGAUA---CGGAUACUUUCCACUUUCGCA ....-........(((.......(((((.---(((((...))))).)).))).....((((((((((((.......---)))))))))))).....))). ( -27.90) >consensus AAAAU___AACGCGCGCAUUGCAUGGGAU_UAUUGGUCUAAUUAAUUCCCCCAAAACGGAAAGUAUCCAAAGUAUCCAGCAGAUACUUUCCACUUUCGCA .............(((........((((....(((((...))))).)))).......((((((((((..............)))))))))).....))). (-15.98 = -16.62 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:18 2006