
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,854,768 – 13,854,873 |
| Length | 105 |
| Max. P | 0.972965 |

| Location | 13,854,768 – 13,854,873 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 77.67 |
| Mean single sequence MFE | -33.37 |
| Consensus MFE | -24.07 |
| Energy contribution | -25.43 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
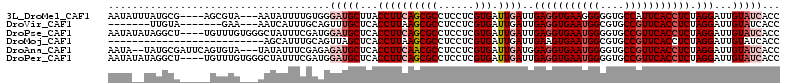
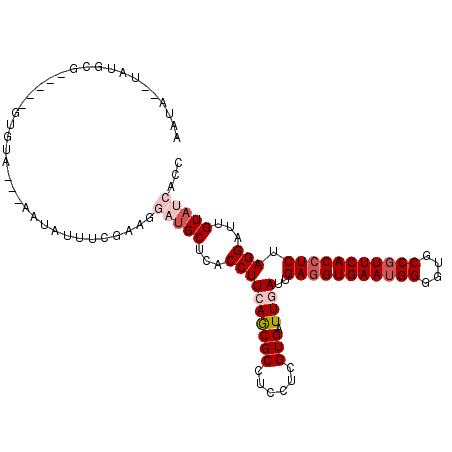
>3L_DroMel_CAF1 13854768 105 + 23771897 AAUAUUUAUGCG----AGCGUA---AAUAUUUUGUGGGAUGCUUACCUUCAGCGCCUCCUCGUGAUUGAUUGAGGUGAAGGGGGUGCCAUUCACCUCUAGGAUUGUAUCACC ((((((((((..----..))))---))))))....(((((((...((((((((((......))).))))..((((((((.((....)).)))))))).)))...))))).)) ( -33.90) >DroVir_CAF1 26435 95 + 1 -------UUGUA-------GAA---AAUCAUUUGCAGUUUGCUCACCUUAAGCGCCUCCUCGUGAUUGAUUGAGGUGAAUGGCGUGCCGUUCACCUCUAGGAUUGUAUCACC -------(((((-------((.---.....)))))))...(((.......)))........(((((((((((((((((((((....)))))))))))...))))).))))). ( -29.00) >DroPse_CAF1 18813 108 + 1 AAUAUAUAGGCU----UGUUUGUGGGCUAUUUCGAUGGAUGCUCACCUUCAGCGCCUCCUCGUGAUUGAUUGAGGUGAAUGGGGUGCCGUUCACCUCUAGGAUUGUAUCACC .......(((((----((...((((((.(((......)))))))))...))).))))....(((((((((((((((((((((....)))))))))))...))))).))))). ( -38.90) >DroMoj_CAF1 21715 86 + 1 --------------------------AGCAUUUGCAGUUAGCUCACCUUAAGCGCCUCCUCGUGAUUGAUUGAAGUGAAUGGCGUGCCGUUCACCUCUAGGAUUGUAUCACC --------------------------.((....)).....(((.......)))........((((((((((((.((((((((....)))))))).))...))))).))))). ( -21.10) >DroAna_CAF1 17536 107 + 1 AAUA--UAUGCGAUUCAGUGUA---UAUAUUUCGAGAGAUGCUCACCUUCAACGCCUCCUCGUGAUUGAUGGAGGUGAAUGGGGUGCCGUUCACCUCUAGGAUUGUAUCACC ((((--((((((......))))---))))))......(((((...((.(((((((......))).))))(((((((((((((....)))))))))))))))...)))))... ( -38.40) >DroPer_CAF1 11122 108 + 1 AAUAUAUAGGCU----UGUUUGUGGGCUAUUUCGAUGGAUGCUCACCUUCAGCGCCUCCUCGUGAUUGAUUGAGGUGAAUGGGGUGCCGUUCACCUCUAGGAUUGUAUCACC .......(((((----((...((((((.(((......)))))))))...))).))))....(((((((((((((((((((((....)))))))))))...))))).))))). ( -38.90) >consensus AAUA__UAUGCG_____GUGUA___AAUAUUUCGAAGGAUGCUCACCUUCAGCGCCUCCUCGUGAUUGAUUGAGGUGAAUGGGGUGCCGUUCACCUCUAGGAUUGUAUCACC .....................................(((((...((((((((((......))).))))..(((((((((((....))))))))))).)))...)))))... (-24.07 = -25.43 + 1.36)

| Location | 13,854,768 – 13,854,873 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 77.67 |
| Mean single sequence MFE | -26.47 |
| Consensus MFE | -19.12 |
| Energy contribution | -19.51 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
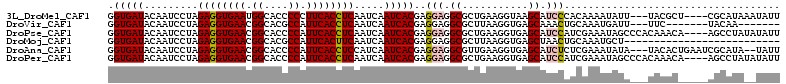
>3L_DroMel_CAF1 13854768 105 - 23771897 GGUGAUACAAUCCUAGAGGUGAAUGGCACCCCCUUCACCUCAAUCAAUCACGAGGAGGCGCUGAAGGUAAGCAUCCCACAAAAUAUU---UACGCU----CGCAUAAAUAUU ((.(((....(((..((((((((.((....)).))))))))..((......)))))...(((.......))))))))....((((((---((.(..----..).)))))))) ( -26.70) >DroVir_CAF1 26435 95 - 1 GGUGAUACAAUCCUAGAGGUGAACGGCACGCCAUUCACCUCAAUCAAUCACGAGGAGGCGCUUAAGGUGAGCAAACUGCAAAUGAUU---UUC-------UACAA------- .(((...........((((((((.((....)).))))))))....(((((...(.((..((((.....))))...)).)...)))))---...-------)))..------- ( -24.30) >DroPse_CAF1 18813 108 - 1 GGUGAUACAAUCCUAGAGGUGAACGGCACCCCAUUCACCUCAAUCAAUCACGAGGAGGCGCUGAAGGUGAGCAUCCAUCGAAAUAGCCCACAAACA----AGCCUAUAUAUU (((.......(((..((((((((.((....)).))))))))..((......)))))((.((((..((((......))))....)))))).......----.)))........ ( -29.40) >DroMoj_CAF1 21715 86 - 1 GGUGAUACAAUCCUAGAGGUGAACGGCACGCCAUUCACUUCAAUCAAUCACGAGGAGGCGCUUAAGGUGAGCUAACUGCAAAUGCU-------------------------- .(((((.........((((((((.((....)).)))))))).....)))))..(.((..((((.....))))...)).).......-------------------------- ( -21.14) >DroAna_CAF1 17536 107 - 1 GGUGAUACAAUCCUAGAGGUGAACGGCACCCCAUUCACCUCCAUCAAUCACGAGGAGGCGUUGAAGGUGAGCAUCUCUCGAAAUAUA---UACACUGAAUCGCAUA--UAUU .(((((.((......((((((((.((....)).)))))))).........((((((.((...........)).)).)))).......---.....)).)))))...--.... ( -27.90) >DroPer_CAF1 11122 108 - 1 GGUGAUACAAUCCUAGAGGUGAACGGCACCCCAUUCACCUCAAUCAAUCACGAGGAGGCGCUGAAGGUGAGCAUCCAUCGAAAUAGCCCACAAACA----AGCCUAUAUAUU (((.......(((..((((((((.((....)).))))))))..((......)))))((.((((..((((......))))....)))))).......----.)))........ ( -29.40) >consensus GGUGAUACAAUCCUAGAGGUGAACGGCACCCCAUUCACCUCAAUCAAUCACGAGGAGGCGCUGAAGGUGAGCAUCCAUCAAAAUAUC___UACAC_____AGCAUA__UAUU .(((((.........((((((((.((....)).)))))))).....)))))..(((.((...........)).))).................................... (-19.12 = -19.51 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:00 2006