
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,833,430 – 13,833,614 |
| Length | 184 |
| Max. P | 0.836720 |

| Location | 13,833,430 – 13,833,538 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.76 |
| Mean single sequence MFE | -24.85 |
| Consensus MFE | -21.17 |
| Energy contribution | -21.42 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.836720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13833430 108 + 23771897 CUCU--------AUAUAUGUAGUAUAUCUAGGUAUAGUACAAGUUCGUGUUGAAUUGUAUGUGU-U---AUAGUGUCUAUAUAGCAGAUACAGAUAUAUAACCAUUUCUUGUUAUGUACU ....--------.....((((.(((((....))))).)))).............((((((.(((-(---(((.......))))))).)))))).((((((((........)))))))).. ( -24.90) >DroSec_CAF1 8366 109 + 1 CUCU--------AUAUAUGUAGUAUAUCUAGGUAUAGUACAAGUUCGUGUUGAAUUGUAUGUGUUU---AUAGUGUCUAUAUAGCAGAUACAGAUAUAUAACCAUUUCUUGUUAUGUACU ....--------.....((((.(((((....))))).)))).............((((((.(((((---((((...))))).)))).)))))).((((((((........)))))))).. ( -23.20) >DroSim_CAF1 7637 109 + 1 CUCU--------AUAUAUGUAGUAUAUCUAGGUAUAGUACAAGUUCGUGUUGAAUUGUAUGUGUUU---AUAGUGUCUAUAUAGCAGAUACAGAUAUAUAACCAUUUCUUGUUAUGUACU ....--------.....((((.(((((....))))).)))).............((((((.(((((---((((...))))).)))).)))))).((((((((........)))))))).. ( -23.20) >DroYak_CAF1 8015 120 + 1 CUCUCUAUACAUAUAUAUGUAGUAUAUCUAGGUAUAGUACAAGUUCGUGUUGAAUUGUAUGUGUUUAGUAUAGUGUCUAUAUAGCAGAUACAGAUAUAUAACCAUUUCUUGUUAUGUACU ....(((.((((((((.((((.(((((....))))).))))(((((.....))))))))))))).)))....((((((.......))))))...((((((((........)))))))).. ( -28.10) >consensus CUCU________AUAUAUGUAGUAUAUCUAGGUAUAGUACAAGUUCGUGUUGAAUUGUAUGUGUUU___AUAGUGUCUAUAUAGCAGAUACAGAUAUAUAACCAUUUCUUGUUAUGUACU .................((((.(((((....))))).)))).............((((((.((((....((((...))))..)))).)))))).((((((((........)))))))).. (-21.17 = -21.42 + 0.25)


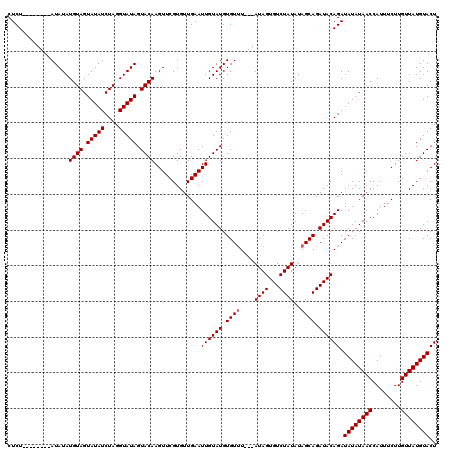
| Location | 13,833,462 – 13,833,578 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.36 |
| Mean single sequence MFE | -11.68 |
| Consensus MFE | -10.10 |
| Energy contribution | -10.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13833462 116 - 23771897 AACCAACAACCAACUACAACAAAAACAAACUGUGCCACCUAGUACAUAACAAGAAAUGGUUAUAUAUCUGUAUCUGCUAUAUAGACACUAU---A-ACACAUACAAUUCAACACGAACUU ..............................(((((......)))))......((((((((((((..(((((((.....)))))))...)))---)-)).)))....)))........... ( -13.70) >DroSec_CAF1 8398 110 - 1 AA-------CCAACUACAACAAAAACAAACUGUGCCACCUAGUACAUAACAAGAAAUGGUUAUAUAUCUGUAUCUGCUAUAUAGACACUAU---AAACACAUACAAUUCAACACGAACUU ..-------.....................((((.....((((..(((((.(....).)))))...(((((((.....))))))).)))).---...))))................... ( -10.90) >DroSim_CAF1 7669 117 - 1 AACCAACAACCAACUACAACAAAAACAAACUGUGCCACCUAGUACAUAACAAGAAAUGGUUAUAUAUCUGUAUCUGCUAUAUAGACACUAU---AAACACAUACAAUUCAACACGAACUU ..............................((((.....((((..(((((.(....).)))))...(((((((.....))))))).)))).---...))))................... ( -10.90) >DroYak_CAF1 8055 120 - 1 AACCAACAACCAACUACAACAAAAACAAACUGUGCCACCUAGUACAUAACAAGAAAUGGUUAUAUAUCUGUAUCUGCUAUAUAGACACUAUACUAAACACAUACAAUUCAACACGAACUU ..............................((((.....(((((.(((((.(....).)))))...(((((((.....))))))).....)))))..))))................... ( -11.20) >consensus AACCAACAACCAACUACAACAAAAACAAACUGUGCCACCUAGUACAUAACAAGAAAUGGUUAUAUAUCUGUAUCUGCUAUAUAGACACUAU___AAACACAUACAAUUCAACACGAACUU ..............................((((.....((((..(((((.(....).)))))...(((((((.....))))))).)))).......))))................... (-10.10 = -10.10 + -0.00)

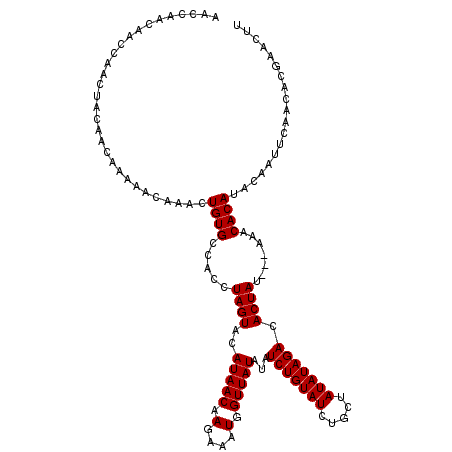
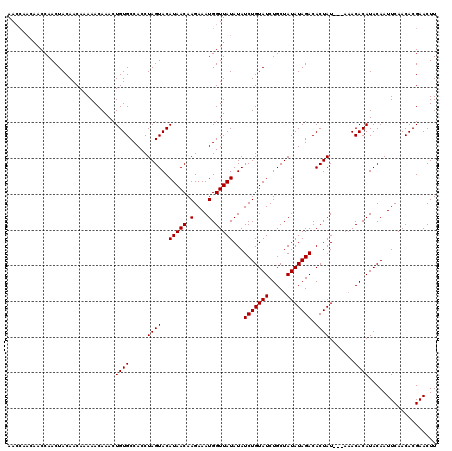
| Location | 13,833,498 – 13,833,614 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 95.69 |
| Mean single sequence MFE | -11.50 |
| Consensus MFE | -11.50 |
| Energy contribution | -11.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13833498 116 - 23771897 AAAACCUUUAAUGAAUUCAGACCAGACCACAAAAACAACCAACAACCAACUACAACAAAAACAAACUGUGCCACCUAGUACAUAACAAGAAAUGGUUAUAUAUCUGUAUCUGCUAU .................((((.((((.........................................((((......))))(((((.(....).)))))...))))..)))).... ( -11.50) >DroSec_CAF1 8435 109 - 1 AAAACCUUUAAUGAAUUCAGACCAGACCACAAAAACAA-------CCAACUACAACAAAAACAAACUGUGCCACCUAGUACAUAACAAGAAAUGGUUAUAUAUCUGUAUCUGCUAU .................((((.((((............-------......................((((......))))(((((.(....).)))))...))))..)))).... ( -11.50) >DroSim_CAF1 7706 116 - 1 AAAACCUUUAAUGAAUUCAGACCAGACCACAAAAACAACCAACAACCAACUACAACAAAAACAAACUGUGCCACCUAGUACAUAACAAGAAAUGGUUAUAUAUCUGUAUCUGCUAU .................((((.((((.........................................((((......))))(((((.(....).)))))...))))..)))).... ( -11.50) >DroYak_CAF1 8095 116 - 1 AAAUCCUUUAAUCAAUUCAGACCAGACCACAAGAACAACCAACAACCAACUACAACAAAAACAAACUGUGCCACCUAGUACAUAACAAGAAAUGGUUAUAUAUCUGUAUCUGCUAU .................((((.((((.........................................((((......))))(((((.(....).)))))...))))..)))).... ( -11.50) >consensus AAAACCUUUAAUGAAUUCAGACCAGACCACAAAAACAACCAACAACCAACUACAACAAAAACAAACUGUGCCACCUAGUACAUAACAAGAAAUGGUUAUAUAUCUGUAUCUGCUAU .................((((.((((.........................................((((......))))(((((.(....).)))))...))))..)))).... (-11.50 = -11.50 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:50 2006