| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,817,656 – 13,817,762 |
| Length | 106 |
| Max. P | 0.878248 |

| Location | 13,817,656 – 13,817,762 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.05 |
| Mean single sequence MFE | -36.51 |
| Consensus MFE | -22.51 |
| Energy contribution | -22.98 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878248 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
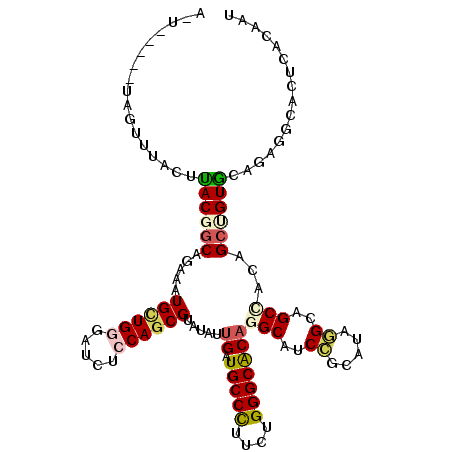
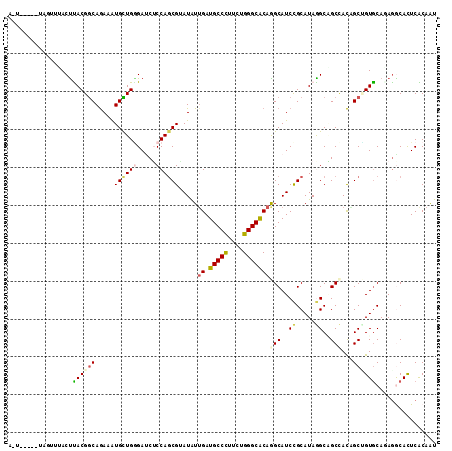
>3L_DroMel_CAF1 13817656 106 - 23771897 A-------UUAUAUACUCACGGCAGAAAUGCUGUGAUCUCCAGCGAAUAUUGAUGCCUUUCUGGGCACAAGCAUCCGCAUAGGCAGCCACAGCAGUGCAGAGACACUCGCAAU .-------........((((((((....))))))))......((((...(((.(((((....))))))))(..((.((((..((.......)).)))).))..)..))))... ( -34.20) >DroVir_CAF1 5803 104 - 1 A-U-----UA---AACCUACGGCAGAAAUGCUGGGAUCGCCAGCGUAUAUUGAUGCCCUUUUGGGCACAGGCAUCCGCAUAGGCCGCCACAGCUGUACACAAGCAUUCACAAU .-.-----..---....((((((....(((((((.....)))))))....((.(((((....)))))))(((..((.....))..)))...))))))................ ( -33.90) >DroGri_CAF1 9559 112 - 1 A-UUAAAAUGGUUUACUUACGACAGAAAUGUUGGGAUCUCCAGCGGAUAUUGAUGCCCUUCUGGGCGCAGGCAUCUGCAUAAGCAGCGACCGCUGUGCAGAGGCAUUCACAAU .-......(((........(((((....)))))......)))(.((((.(((.(((((....))))))))((.(((((....(((((....)))))))))).)))))).)... ( -36.74) >DroEre_CAF1 7490 110 - 1 UACUU---AAGUGAACUCACGGCAAAAGUGCUGGGACCUCCAGCGAAUAUUGAUGCCCUUCUGGGCACAGGCAUCCGCAUAGGCAGCCAUGGCAGUGCAGAGGCACUCGCAAU .....---..(((....)))(((....(((((((.....))))).....(((.(((((....))))))))))....((....)).)))...(((((((....))))).))... ( -37.70) >DroMoj_CAF1 5925 107 - 1 A-U-----UCAUCGACUUACGGCAGAAAUGUUGGGAUCGCCAGCGUAUGUUGAUGCCCUUCUGGGCACAGGCAUCCGCAUAGGCCGCCACAGCCGUGCACAGGCACUCGCAAU .-.-----...........(((((....)))))((...(((.(((.(((((..(((((....)))))..))))).)))...)))..))...((.((((....))))..))... ( -38.40) >DroAna_CAF1 126320 105 - 1 --------AGGUUUACUUACGGCAGAAGUGCUGAGACCUCCAUCGUAUGUGGAUGCCCUUCUGGGCACAGGCAUCCGCAUAAGCGGCUACUGCUGUGCAAAGACACUCACAGU --------..((((...((((((((.((((((((........)).((((((((((((....((....))))))))))))))))).))).))))))))...))))......... ( -38.10) >consensus A_U_____UAGUUUACUUACGGCAGAAAUGCUGGGAUCUCCAGCGUAUAUUGAUGCCCUUCUGGGCACAGGCAUCCGCAUAGGCAGCCACAGCUGUGCAGAGGCACUCACAAU .................((((((.....((((((.....)))))).....((.(((((....)))))))(((..((.....))..)))...))))))................ (-22.51 = -22.98 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:45 2006