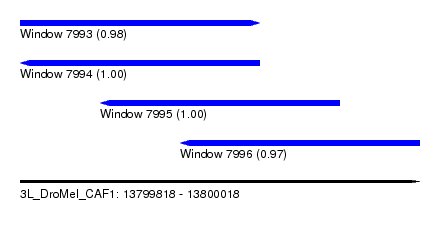
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,799,818 – 13,800,018 |
| Length | 200 |
| Max. P | 0.999823 |

| Location | 13,799,818 – 13,799,938 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -29.44 |
| Consensus MFE | -28.30 |
| Energy contribution | -28.14 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13799818 120 + 23771897 ACUCAAGUGAAUUAAAUUGGUUUCCAGCUGGUCUGUGCUAUUAGUUGUAAUUGAUUCUUGAAAUAAUUUAUUUUAUGGCUUUUUGUCGCCAACCGCCUCCGGCUGACGAUUGACUUCACU .....((((((...(((((.....(((((((...(.((....................(((((((...)))))))((((........))))...))).))))))).)))))...)))))) ( -27.70) >DroSec_CAF1 36648 120 + 1 ACUCAAGUGAAUUAAAUUGGUUUCCAGCUGGUCUGUGCUAUUAGUUGUAAUUGAUUCUUGAAAUAAUUUAUUUCAUGGCUUUUUGUCGCCAACCGCCUCCGGCUGACGAUUGCCUUCACU .....((((((...(((..(((..((((((((.......)))))))).)))..)))..(((((((...))))))).(((...((((((((..........))).)))))..))))))))) ( -30.30) >DroSim_CAF1 32834 120 + 1 ACUCAAGUGAAUUAAAUUGGUUUCCAGCUGGUCUGUGCUAUUAGUUGUAAUUGAUUCUUGAAAUAAUUUAUUUUAUGGCUUUUUGUCGCCAACCGCCUCCGGCUGACGAUUGCCUUCACU .....((((((...(((..(((..((((((((.......)))))))).)))..)))..(((((((...))))))).(((...((((((((..........))).)))))..))))))))) ( -28.10) >DroEre_CAF1 37523 120 + 1 ACUCAAGUGAAUUAAAUUGGUUUCCAGCUGGUCUGUGCUAUUAGUUGUAAUUGAUUCUUGAAAUAAUUUAUUUUAUGGCUUUUUGUCGCCAGCCGCCUCCGGCUGACGAUUGCCUUCACU .....((((((...(((..(((..((((((((.......)))))))).)))..)))..(((((((...))))))).(((.....((((.((((((....)))))).)))).))))))))) ( -33.00) >DroYak_CAF1 35355 120 + 1 ACUCAAGUGAAUUAAAUUGGUUUCCAGCUGGUCUGUGCUAUUAGUUGUAAUUGAUUCUUGAAAUAAUUUAUUUUAUGGCUUUUUGUCGCCAACCGCCUCCGGCUGACGAUUGCCUUCACU .....((((((...(((..(((..((((((((.......)))))))).)))..)))..(((((((...))))))).(((...((((((((..........))).)))))..))))))))) ( -28.10) >consensus ACUCAAGUGAAUUAAAUUGGUUUCCAGCUGGUCUGUGCUAUUAGUUGUAAUUGAUUCUUGAAAUAAUUUAUUUUAUGGCUUUUUGUCGCCAACCGCCUCCGGCUGACGAUUGCCUUCACU .....((((((...(((((.....(((((((...(.((....................(((((((...)))))))((((........))))...))).))))))).)))))...)))))) (-28.30 = -28.14 + -0.16)

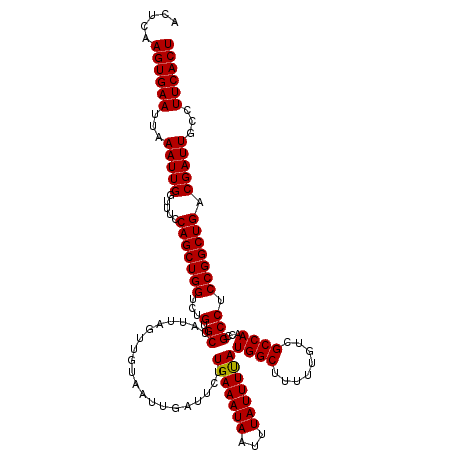
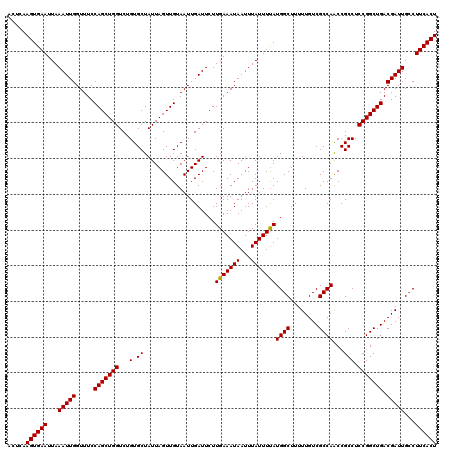
| Location | 13,799,818 – 13,799,938 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -28.56 |
| Consensus MFE | -27.50 |
| Energy contribution | -27.54 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.998388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13799818 120 - 23771897 AGUGAAGUCAAUCGUCAGCCGGAGGCGGUUGGCGACAAAAAGCCAUAAAAUAAAUUAUUUCAAGAAUCAAUUACAACUAAUAGCACAGACCAGCUGGAAACCAAUUUAAUUCACUUGAGU ((((((.....((((((((((....))))))))))...............((((((.((((.........(((....)))((((........))))))))..)))))).))))))..... ( -25.10) >DroSec_CAF1 36648 120 - 1 AGUGAAGGCAAUCGUCAGCCGGAGGCGGUUGGCGACAAAAAGCCAUGAAAUAAAUUAUUUCAAGAAUCAAUUACAACUAAUAGCACAGACCAGCUGGAAACCAAUUUAAUUCACUUGAGU (((((((((..((((((((((....))))))))))......))).(((((((...)))))))..................((((........)))).............))))))..... ( -30.10) >DroSim_CAF1 32834 120 - 1 AGUGAAGGCAAUCGUCAGCCGGAGGCGGUUGGCGACAAAAAGCCAUAAAAUAAAUUAUUUCAAGAAUCAAUUACAACUAAUAGCACAGACCAGCUGGAAACCAAUUUAAUUCACUUGAGU (((((((((..((((((((((....))))))))))......)))......((((((.((((.........(((....)))((((........))))))))..)))))).))))))..... ( -28.40) >DroEre_CAF1 37523 120 - 1 AGUGAAGGCAAUCGUCAGCCGGAGGCGGCUGGCGACAAAAAGCCAUAAAAUAAAUUAUUUCAAGAAUCAAUUACAACUAAUAGCACAGACCAGCUGGAAACCAAUUUAAUUCACUUGAGU (((((((((..((((((((((....))))))))))......)))......((((((.((((.........(((....)))((((........))))))))..)))))).))))))..... ( -30.80) >DroYak_CAF1 35355 120 - 1 AGUGAAGGCAAUCGUCAGCCGGAGGCGGUUGGCGACAAAAAGCCAUAAAAUAAAUUAUUUCAAGAAUCAAUUACAACUAAUAGCACAGACCAGCUGGAAACCAAUUUAAUUCACUUGAGU (((((((((..((((((((((....))))))))))......)))......((((((.((((.........(((....)))((((........))))))))..)))))).))))))..... ( -28.40) >consensus AGUGAAGGCAAUCGUCAGCCGGAGGCGGUUGGCGACAAAAAGCCAUAAAAUAAAUUAUUUCAAGAAUCAAUUACAACUAAUAGCACAGACCAGCUGGAAACCAAUUUAAUUCACUUGAGU (((((((((..((((((((((....))))))))))......)))......((((((.((((.........(((....)))((((........))))))))..)))))).))))))..... (-27.50 = -27.54 + 0.04)

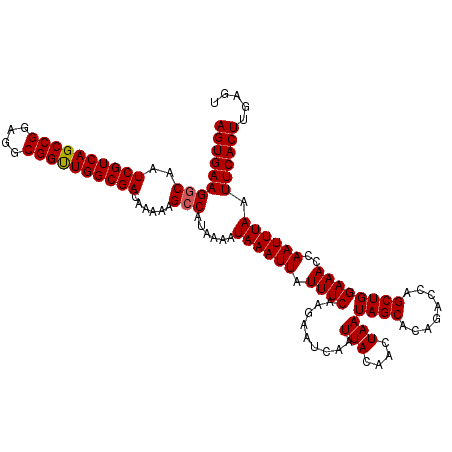
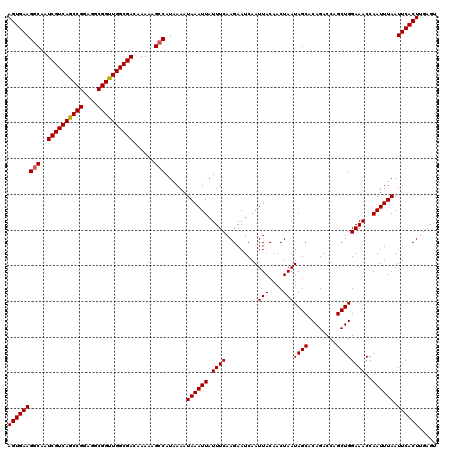
| Location | 13,799,858 – 13,799,978 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -28.90 |
| Energy contribution | -28.94 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.17 |
| SVM RNA-class probability | 0.999823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13799858 120 - 23771897 GCAAUUUGCCGAACUUUCAACUCCACUCCAAGGAUCGUUAAGUGAAGUCAAUCGUCAGCCGGAGGCGGUUGGCGACAAAAAGCCAUAAAAUAAAUUAUUUCAAGAAUCAAUUACAACUAA ((...(((.(..((((.....(((.......))).....))))...).)))((((((((((....))))))))))......))..................................... ( -22.70) >DroSec_CAF1 36688 120 - 1 GCAAUUUGCCGAACUUUCAACUCCACUCCUAGGAUCGUUAAGUGAAGGCAAUCGUCAGCCGGAGGCGGUUGGCGACAAAAAGCCAUGAAAUAAAUUAUUUCAAGAAUCAAUUACAACUAA ((...(((((..((((.....(((.......))).....))))...)))))((((((((((....))))))))))......))..(((((((...))))))).................. ( -32.90) >DroSim_CAF1 32874 120 - 1 GCAAUUUGCCGAACUUUCAACUCCACUCCAAGGAUCGUUAAGUGAAGGCAAUCGUCAGCCGGAGGCGGUUGGCGACAAAAAGCCAUAAAAUAAAUUAUUUCAAGAAUCAAUUACAACUAA ((...(((((..((((.....(((.......))).....))))...)))))((((((((((....))))))))))......))..................................... ( -29.80) >DroEre_CAF1 37563 119 - 1 GCAAUUUGCCGAACUUUCAACUCCACU-AAAGGAUCGUUAAGUGAAGGCAAUCGUCAGCCGGAGGCGGCUGGCGACAAAAAGCCAUAAAAUAAAUUAUUUCAAGAAUCAAUUACAACUAA ((...(((((..((((.....(((...-...))).....))))...)))))((((((((((....))))))))))......))..................................... ( -32.70) >DroYak_CAF1 35395 119 - 1 GCAAUUUGCCGAACUUUCAACUCCACC-AAAGGACCGUUAAGUGAAGGCAAUCGUCAGCCGGAGGCGGUUGGCGACAAAAAGCCAUAAAAUAAAUUAUUUCAAGAAUCAAUUACAACUAA ((...(((((..((((.....(((...-...))).....))))...)))))((((((((((....))))))))))......))..................................... ( -30.30) >consensus GCAAUUUGCCGAACUUUCAACUCCACUCCAAGGAUCGUUAAGUGAAGGCAAUCGUCAGCCGGAGGCGGUUGGCGACAAAAAGCCAUAAAAUAAAUUAUUUCAAGAAUCAAUUACAACUAA ((...(((((..((((.....(((.......))).....))))...)))))((((((((((....))))))))))......))..................................... (-28.90 = -28.94 + 0.04)

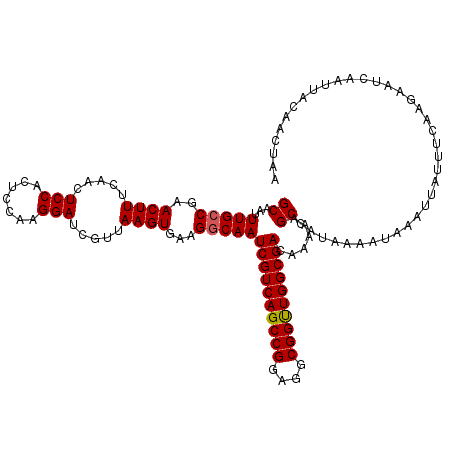
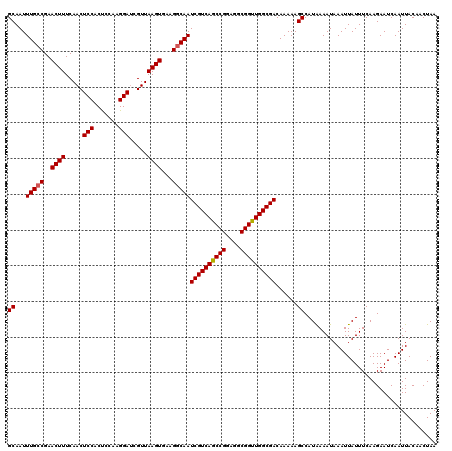
| Location | 13,799,898 – 13,800,018 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -32.68 |
| Consensus MFE | -30.90 |
| Energy contribution | -30.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13799898 120 - 23771897 AAUUCAAGCCAUUGACAUUGACAGGAAAUCCCGAUUAUAUGCAAUUUGCCGAACUUUCAACUCCACUCCAAGGAUCGUUAAGUGAAGUCAAUCGUCAGCCGGAGGCGGUUGGCGACAAAA ...((((....))))..(((((.((.....))...........(((((.(((.((((............)))).))).)))))...)))))((((((((((....))))))))))..... ( -27.30) >DroSec_CAF1 36728 120 - 1 AAUUUAAGCCAUUGACAUUGACAGGAAAUCCCGAUUAUGUGCAAUUUGCCGAACUUUCAACUCCACUCCUAGGAUCGUUAAGUGAAGGCAAUCGUCAGCCGGAGGCGGUUGGCGACAAAA ..........((((((((...(.((....)).)...)))).))))(((((..((((.....(((.......))).....))))...)))))((((((((((....))))))))))..... ( -33.70) >DroSim_CAF1 32914 120 - 1 AAUUUAAGCCAUUGAAAUUGACAGGAAAUCCCGAUUAUGUGCAAUUUGCCGAACUUUCAACUCCACUCCAAGGAUCGUUAAGUGAAGGCAAUCGUCAGCCGGAGGCGGUUGGCGACAAAA .......(((((...(((((...((....)))))))))).))...(((((..((((.....(((.......))).....))))...)))))((((((((((....))))))))))..... ( -33.40) >DroEre_CAF1 37603 119 - 1 AAUUUAAGCCAUUGACAUUGACAGGAAAUCCCGAUUAUAUGCAAUUUGCCGAACUUUCAACUCCACU-AAAGGAUCGUUAAGUGAAGGCAAUCGUCAGCCGGAGGCGGCUGGCGACAAAA ..........((((.((((((..((.....))..))).)))))))(((((..((((.....(((...-...))).....))))...)))))((((((((((....))))))))))..... ( -35.80) >DroYak_CAF1 35435 119 - 1 AAUUUAAGCCAUUGACAUUGACAGGAAAUGCCGAUUAUAUGCAAUUUGCCGAACUUUCAACUCCACC-AAAGGACCGUUAAGUGAAGGCAAUCGUCAGCCGGAGGCGGUUGGCGACAAAA ..........((((.((((((..((.....))..))).)))))))(((((..((((.....(((...-...))).....))))...)))))((((((((((....))))))))))..... ( -33.20) >consensus AAUUUAAGCCAUUGACAUUGACAGGAAAUCCCGAUUAUAUGCAAUUUGCCGAACUUUCAACUCCACUCCAAGGAUCGUUAAGUGAAGGCAAUCGUCAGCCGGAGGCGGUUGGCGACAAAA ..........((((.((((((..((.....))..))).)))))))(((((..((((.....(((.......))).....))))...)))))((((((((((....))))))))))..... (-30.90 = -30.90 + 0.00)

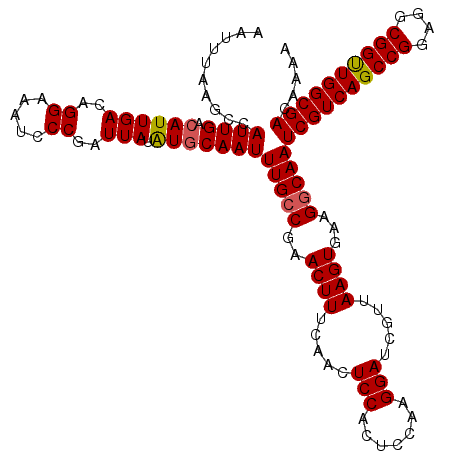
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:37 2006