| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,475,478 – 1,475,577 |
| Length | 99 |
| Max. P | 0.500000 |

| Location | 1,475,478 – 1,475,577 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 76.76 |
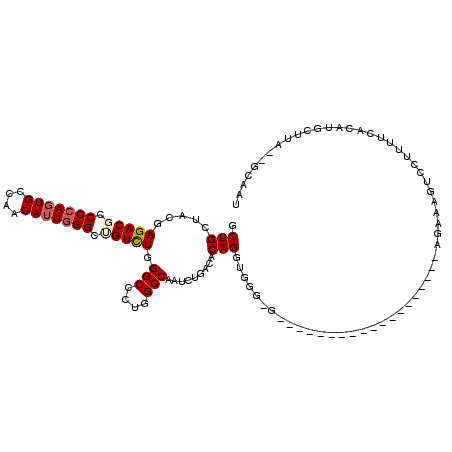
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -20.68 |
| Energy contribution | -21.02 |
| Covariance contribution | 0.33 |
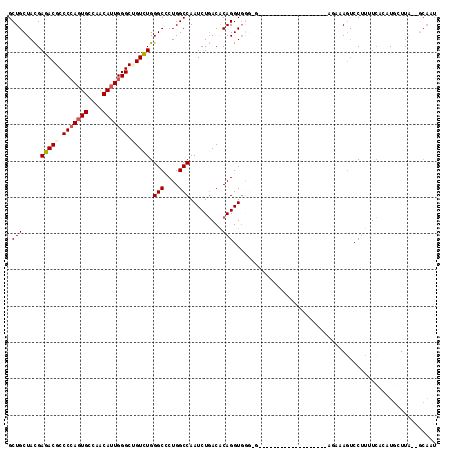
| Combinations/Pair | 1.11 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
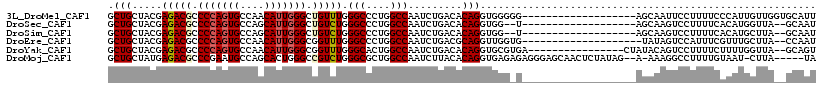
>3L_DroMel_CAF1 1475478 99 - 23771897 GCUGCUACGAGACGCCCCAGUGCCAACAUUGGGCUGUUUGGGCCCUGGCCAAUCUGACACAGGUGGGGG-------------------AGCAAUUCCUUUUCCCAUUGUUGGUGCAUU ((((((.(.(((((.(((((((....))))))).))))).).((((.(((...........))).))))-------------------))))..........(((....))).))... ( -34.10) >DroSec_CAF1 122427 95 - 1 GCUGCUACGAGACGCCCCAGUGCCAGCAUUGGGCUGUCUGGGCCCUGGCCAAUCUGACACAGGUGG--U-------------------AGCAAGUCCUUUUCACAUGGUUA--GCAAU ((((((((.(((((.(((((((....))))))).))))).(((....)))............))))--)-------------------)))....................--..... ( -36.00) >DroSim_CAF1 119874 95 - 1 GCUGCUACGAGACGCCCCAGUGCCAGCAUUGGGCUGUCUGGGCCCUGGCCAAUCUGACACAGGUGG--U-------------------AGCAAGUCCUUUUCACAUGCUUA--GCAAU ((((((((.(((((.(((((((....))))))).))))).(((....)))............))))--)-------------------)))..............(((...--))).. ( -36.40) >DroEre_CAF1 127179 96 - 1 GCUGCUACGAGACGCCCCAGUGCCAACAUUGGGCGGUUUGGGCCCUGGCCAAUCUGACGCAGGUUGGUG--------------------UAUAGUCCAUUUCGUUUGCUUA--CCAAU ...((.(((((((((((.((((....)))))))))...(((((....(((((((((...))))))))).--------------------....)))))))))))..))...--..... ( -35.60) >DroYak_CAF1 128024 100 - 1 GCUGCUACGAGACGCCCCAGUGCCAACAUUGGGCGGUUUGGGCACUGGCCAAUCUGACACAGGUGCGUGA----------------CUAUACAGUCCUUUUCUUUUGGUUA--GCAGU ((((((((.((((((.((.(((.((..(((((.((((......)))).))))).)).))).)).))))((----------------((....))))........)).).))--))))) ( -33.60) >DroMoj_CAF1 109780 109 - 1 GCUGCUAUGAGACGCCCGAAUGCCAGCACUGGGCCGUCUGGGCGCUGGCCAAUCUUACACAGGUGAGAGAGGGAGCAACUCUAUAG--A-AAAGGCCUUUUGUAAU-CUUA-----UA .....((((((((((((((..(((.......)))..)).)))))..((((..((((((....))))))(((.......))).....--.-...))))........)-))))-----)) ( -35.50) >consensus GCUGCUACGAGACGCCCCAGUGCCAACAUUGGGCUGUCUGGGCCCUGGCCAAUCUGACACAGGUGGG_G___________________AGAAAGUCCUUUUCACAUGCUUA__GCAAU .(((.....(((((.(((((((....))))))).))))).(((....))).........)))........................................................ (-20.68 = -21.02 + 0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:42 2006