
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,799,393 – 13,799,671 |
| Length | 278 |
| Max. P | 0.999932 |

| Location | 13,799,393 – 13,799,513 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.42 |
| Mean single sequence MFE | -21.04 |
| Consensus MFE | -17.98 |
| Energy contribution | -18.58 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13799393 120 + 23771897 UUAUUAAGCUGCCAAAUCAAAUUUUAAACUUCCCAGCUCAAAAGGCCCACUUGACCAAUCUACCAAGGAAAGCUUUCAAAGAUCCCUUUCAUUGCUCCCCGGGCCACAUCCAAACCGGCU ..........((((((......)))..................(((((..(((..........)))((..(((..(.((((....)))).)..)))..)))))))...........))). ( -21.60) >DroSec_CAF1 36235 120 + 1 UUAUUAACCUGCCAAAUCAAAUUUUAAACUUCCCAGUUCAAAAGGCCCACUUGACCAAUCUACAAAGGAAAGCUUUCAAAGAUCCCUUUCAUUGCUCCCCGGGCCACAUCCAAACCGGCU ..........(((........((((.((((....)))).))))(((((....((....))......((..(((..(.((((....)))).)..)))..)))))))...........))). ( -24.60) >DroSim_CAF1 32421 120 + 1 UCAUUAACCUGCCAAAUCAAAUUUUAAACUUCCCAGCUCAAAAGGCCCACUUGACCAAUCUACAAAGGAAAGCUUUCAAAGAUCCCUUUCAUUGCUCCCCGGGCCACAUCCAAGCCGGCU ..........((((((......)))..........(((.....(((((....((....))......((..(((..(.((((....)))).)..)))..))))))).......))).))). ( -21.50) >DroEre_CAF1 37126 120 + 1 UUAUAAACCUGCCAAAUCAAAUUUUAAACUUCGCAGCUCAAAAGGCCCACUUGACCAAUCUGCAAAGGAAAGCUUUCAAAGAUCCCAUCCAUUGCUCCCCGGGCCACAUUCAAACUGGCU ..........((((..............((....)).......(((((....((....)).((((.(((...((.....))......))).)))).....)))))..........)))). ( -19.50) >DroYak_CAF1 34932 120 + 1 AUAUAAACCUGCCAAAUCAAAUUUGAAACUUCCCAGCUGAAAAGACCCACUUGACCAAUCUACAAAGGAUAGCUUUCAAAGAUCCCUUUCAUUGCUCCCCGGGCCACUUGCAAACUGGCU ..........((((..(((....))).....(((..((....))......................((..(((..(.((((....)))).)..)))..)))))............)))). ( -18.00) >consensus UUAUUAACCUGCCAAAUCAAAUUUUAAACUUCCCAGCUCAAAAGGCCCACUUGACCAAUCUACAAAGGAAAGCUUUCAAAGAUCCCUUUCAUUGCUCCCCGGGCCACAUCCAAACCGGCU ..........(((...............((....)).......(((((....((....))......((..(((..(.((((....)))).)..)))..)))))))...........))). (-17.98 = -18.58 + 0.60)



| Location | 13,799,393 – 13,799,513 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.42 |
| Mean single sequence MFE | -36.72 |
| Consensus MFE | -33.98 |
| Energy contribution | -33.94 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.69 |
| SVM RNA-class probability | 0.999532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13799393 120 - 23771897 AGCCGGUUUGGAUGUGGCCCGGGGAGCAAUGAAAGGGAUCUUUGAAAGCUUUCCUUGGUAGAUUGGUCAAGUGGGCCUUUUGAGCUGGGAAGUUUAAAAUUUGAUUUGGCAGCUUAAUAA .(((((.(..(((..(((((((..(((..(.((((....)))).)..)))..))((((........))))..))))).((((((((....)))))))))))..).))))).......... ( -38.60) >DroSec_CAF1 36235 120 - 1 AGCCGGUUUGGAUGUGGCCCGGGGAGCAAUGAAAGGGAUCUUUGAAAGCUUUCCUUUGUAGAUUGGUCAAGUGGGCCUUUUGAACUGGGAAGUUUAAAAUUUGAUUUGGCAGGUUAAUAA .(((((.(..(((..(((((((..(((..(.((((....)))).)..)))..))......((....))....))))).((((((((....)))))))))))..).))))).......... ( -38.20) >DroSim_CAF1 32421 120 - 1 AGCCGGCUUGGAUGUGGCCCGGGGAGCAAUGAAAGGGAUCUUUGAAAGCUUUCCUUUGUAGAUUGGUCAAGUGGGCCUUUUGAGCUGGGAAGUUUAAAAUUUGAUUUGGCAGGUUAAUGA ..((((((..((...(((((((..(((..(.((((....)))).)..)))..))......((....))....))))).))..)))))).........((((((......))))))..... ( -39.30) >DroEre_CAF1 37126 120 - 1 AGCCAGUUUGAAUGUGGCCCGGGGAGCAAUGGAUGGGAUCUUUGAAAGCUUUCCUUUGCAGAUUGGUCAAGUGGGCCUUUUGAGCUGCGAAGUUUAAAAUUUGAUUUGGCAGGUUUAUAA .(((((.(..(((..((((((..((.((((.(..((((.............))))...)..)))).))...)))))).((((((((....)))))))))))..).))))).......... ( -36.82) >DroYak_CAF1 34932 120 - 1 AGCCAGUUUGCAAGUGGCCCGGGGAGCAAUGAAAGGGAUCUUUGAAAGCUAUCCUUUGUAGAUUGGUCAAGUGGGUCUUUUCAGCUGGGAAGUUUCAAAUUUGAUUUGGCAGGUUUAUAU .(((((....(((((((((((..((.((((..(((((((((.....))..)))))))....)))).))...))))))((..(....)..)).......)))))..))))).......... ( -30.70) >consensus AGCCGGUUUGGAUGUGGCCCGGGGAGCAAUGAAAGGGAUCUUUGAAAGCUUUCCUUUGUAGAUUGGUCAAGUGGGCCUUUUGAGCUGGGAAGUUUAAAAUUUGAUUUGGCAGGUUAAUAA .(((((.(..((...(((((((..(((..(.((((....)))).)..)))..))......((....))....)))))(((((((((....)))))))))))..).))))).......... (-33.98 = -33.94 + -0.04)



| Location | 13,799,473 – 13,799,591 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.55 |
| Mean single sequence MFE | -33.72 |
| Consensus MFE | -30.08 |
| Energy contribution | -29.68 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13799473 118 + 23771897 GAUCCCUUUCAUUGCUCCCCGGGCCACAUCCAAACCGGCUAAUUUCCAAAGACUGGAUGCUGU--GUGCUUGGCCUUGUGCUUAUGCAAACCGAAAUCGGCUUGCCAAGCAUUACAAUCA (((..........((...((((............))))......((((.....)))).))(((--(((((((((....(((....)))..(((....)))...)))))))).))))))). ( -32.40) >DroSec_CAF1 36315 118 + 1 GAUCCCUUUCAUUGCUCCCCGGGCCACAUCCAAACCGGCUAAUUUCCGAAGACUGGAUGCUGU--GUGCUUGGCCUUGUGCUUAUGCAAACCGAAAUCGGCUUGCCAAGCAUUACAAUCA (((..........(((.....)))..((((((...(((.......))).....)))))).(((--(((((((((....(((....)))..(((....)))...)))))))).))))))). ( -33.00) >DroSim_CAF1 32501 118 + 1 GAUCCCUUUCAUUGCUCCCCGGGCCACAUCCAAGCCGGCUAAUUUCCGAAGACUGGAUGCUGU--GUGCUUGGCCUUGUGCUUAUGCAAACCGAAAUCGGCUUGCCAAGCAUUACAAUCA (((..........(((.....)))..((((((..((((.......)))..)..)))))).(((--(((((((((....(((....)))..(((....)))...)))))))).))))))). ( -33.60) >DroEre_CAF1 37206 120 + 1 GAUCCCAUCCAUUGCUCCCCGGGCCACAUUCAAACUGGCUAAUUUCCGAAGACUGGAUGCUGUGUGUGCUUGGCCUUGUGCUUAUGCAAACCGAAAUCGGCUUGCCAAGCAUUACAAUCA (((..((((((...((...(((((((.........))))......))).))..)))))).((((.(((((((((....(((....)))..(((....)))...)))))))))))))))). ( -35.70) >DroYak_CAF1 35012 118 + 1 GAUCCCUUUCAUUGCUCCCCGGGCCACUUGCAAACUGGCUAAUUUCCGAAGACUGGAUGCUGU--GUGCUUGGCCUUGUGCUUAUGCAAACCGAAAUCGGCUUGCCAAGCAUUACAAUCA (((.................((((((...(((.((.(((.....((((.....)))).)))))--.))).)))))).((((((..((((.(((....))).)))).))))))....))). ( -33.90) >consensus GAUCCCUUUCAUUGCUCCCCGGGCCACAUCCAAACCGGCUAAUUUCCGAAGACUGGAUGCUGU__GUGCUUGGCCUUGUGCUUAUGCAAACCGAAAUCGGCUUGCCAAGCAUUACAAUCA (((.................((((((....((...((((.....((((.....)))).))))....))..)))))).((((((..((((.(((....))).)))).))))))....))). (-30.08 = -29.68 + -0.40)


| Location | 13,799,473 – 13,799,591 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.55 |
| Mean single sequence MFE | -42.24 |
| Consensus MFE | -37.88 |
| Energy contribution | -38.64 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.12 |
| SVM RNA-class probability | 0.998505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13799473 118 - 23771897 UGAUUGUAAUGCUUGGCAAGCCGAUUUCGGUUUGCAUAAGCACAAGGCCAAGCAC--ACAGCAUCCAGUCUUUGGAAAUUAGCCGGUUUGGAUGUGGCCCGGGGAGCAAUGAAAGGGAUC (.(((((..((((((((((((((....)))))))).))))))(..(((((.((..--...))((((((...((((.......)))).)))))).)))))..)...))))).)........ ( -41.70) >DroSec_CAF1 36315 118 - 1 UGAUUGUAAUGCUUGGCAAGCCGAUUUCGGUUUGCAUAAGCACAAGGCCAAGCAC--ACAGCAUCCAGUCUUCGGAAAUUAGCCGGUUUGGAUGUGGCCCGGGGAGCAAUGAAAGGGAUC (.(((((..((((((((((((((....)))))))).))))))(..(((((.((..--...))((((((...((((.......)))).)))))).)))))..)...))))).)........ ( -43.70) >DroSim_CAF1 32501 118 - 1 UGAUUGUAAUGCUUGGCAAGCCGAUUUCGGUUUGCAUAAGCACAAGGCCAAGCAC--ACAGCAUCCAGUCUUCGGAAAUUAGCCGGCUUGGAUGUGGCCCGGGGAGCAAUGAAAGGGAUC (.(((((..((((((((((((((....)))))))).))))))(..(((((.((..--...))((((((...((((.......)))).)))))).)))))..)...))))).)........ ( -42.30) >DroEre_CAF1 37206 120 - 1 UGAUUGUAAUGCUUGGCAAGCCGAUUUCGGUUUGCAUAAGCACAAGGCCAAGCACACACAGCAUCCAGUCUUCGGAAAUUAGCCAGUUUGAAUGUGGCCCGGGGAGCAAUGGAUGGGAUC ...((((..((((((((((((((....))))))((....)).....))))))))...))))((((((.(((((((......((((.........)))))))))))....))))))..... ( -42.90) >DroYak_CAF1 35012 118 - 1 UGAUUGUAAUGCUUGGCAAGCCGAUUUCGGUUUGCAUAAGCACAAGGCCAAGCAC--ACAGCAUCCAGUCUUCGGAAAUUAGCCAGUUUGCAAGUGGCCCGGGGAGCAAUGAAAGGGAUC (.(((((..((((((((((((((....)))))))).))))))(..(((((.(((.--((.((......((....)).....))..)).)))...)))))..)...))))).)........ ( -40.60) >consensus UGAUUGUAAUGCUUGGCAAGCCGAUUUCGGUUUGCAUAAGCACAAGGCCAAGCAC__ACAGCAUCCAGUCUUCGGAAAUUAGCCGGUUUGGAUGUGGCCCGGGGAGCAAUGAAAGGGAUC (.(((((..((((((((((((((....)))))))).))))))(..((((..(......).((((((((...((((.......)))).))))))))))))..)...))))).)........ (-37.88 = -38.64 + 0.76)


| Location | 13,799,513 – 13,799,631 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.48 |
| Mean single sequence MFE | -33.14 |
| Consensus MFE | -31.74 |
| Energy contribution | -31.58 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13799513 118 + 23771897 AAUUUCCAAAGACUGGAUGCUGU--GUGCUUGGCCUUGUGCUUAUGCAAACCGAAAUCGGCUUGCCAAGCAUUACAAUCAAUUAUGCAUUGGCUUAAAUCAUCGCAUAUGUGAUUUACAU ....((((.....))))....((--(((((((((....(((....)))..(((....)))...)))))))).)))(((((..(((((..(((......)))..)))))..)))))..... ( -33.60) >DroSec_CAF1 36355 118 + 1 AAUUUCCGAAGACUGGAUGCUGU--GUGCUUGGCCUUGUGCUUAUGCAAACCGAAAUCGGCUUGCCAAGCAUUACAAUCAAUUAUGCAUUGGCUUAAAUCAUCGCAUAUGUGAUUUACAU ....((((.....))))....((--(((((((((....(((....)))..(((....)))...)))))))).)))(((((..(((((..(((......)))..)))))..)))))..... ( -32.90) >DroSim_CAF1 32541 118 + 1 AAUUUCCGAAGACUGGAUGCUGU--GUGCUUGGCCUUGUGCUUAUGCAAACCGAAAUCGGCUUGCCAAGCAUUACAAUCAAUUAUGCAUUGGCUUAAAUCAUCGCAUAUGUGAUUUACAU ....((((.....))))....((--(((((((((....(((....)))..(((....)))...)))))))).)))(((((..(((((..(((......)))..)))))..)))))..... ( -32.90) >DroEre_CAF1 37246 120 + 1 AAUUUCCGAAGACUGGAUGCUGUGUGUGCUUGGCCUUGUGCUUAUGCAAACCGAAAUCGGCUUGCCAAGCAUUACAAUCAAUUAUGCAUUGGCUUAAAUCAUCGCAUACGUGAUUUACAU ....((((.....))))....(((.(((((((((....(((....)))..(((....)))...))))))))))))(((((..(((((..(((......)))..)))))..)))))..... ( -33.40) >DroYak_CAF1 35052 118 + 1 AAUUUCCGAAGACUGGAUGCUGU--GUGCUUGGCCUUGUGCUUAUGCAAACCGAAAUCGGCUUGCCAAGCAUUACAAUCAAUUAUGCAUUGGCUUAAAUCAUCGCAUACGUGAUUUACAU ....((((.....))))....((--(((((((((....(((....)))..(((....)))...)))))))).)))(((((..(((((..(((......)))..)))))..)))))..... ( -32.90) >consensus AAUUUCCGAAGACUGGAUGCUGU__GUGCUUGGCCUUGUGCUUAUGCAAACCGAAAUCGGCUUGCCAAGCAUUACAAUCAAUUAUGCAUUGGCUUAAAUCAUCGCAUAUGUGAUUUACAU ....((((.....))))........(((((((((....(((....)))..(((....)))...)))))))))...(((((..(((((..(((......)))..)))))..)))))..... (-31.74 = -31.58 + -0.16)


| Location | 13,799,513 – 13,799,631 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.48 |
| Mean single sequence MFE | -35.70 |
| Consensus MFE | -35.58 |
| Energy contribution | -35.58 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.26 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.64 |
| SVM RNA-class probability | 0.999932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13799513 118 - 23771897 AUGUAAAUCACAUAUGCGAUGAUUUAAGCCAAUGCAUAAUUGAUUGUAAUGCUUGGCAAGCCGAUUUCGGUUUGCAUAAGCACAAGGCCAAGCAC--ACAGCAUCCAGUCUUUGGAAAUU .(((.(((((..((((((.((........)).))))))..)))))....((((((((((((((....))))))((....)).....)))))))).--...)))(((((...))))).... ( -35.60) >DroSec_CAF1 36355 118 - 1 AUGUAAAUCACAUAUGCGAUGAUUUAAGCCAAUGCAUAAUUGAUUGUAAUGCUUGGCAAGCCGAUUUCGGUUUGCAUAAGCACAAGGCCAAGCAC--ACAGCAUCCAGUCUUCGGAAAUU ((((.(((((..((((((.((........)).))))))..)))))....((((((((((((((....))))))((....)).....)))))))).--...))))....((....)).... ( -35.20) >DroSim_CAF1 32541 118 - 1 AUGUAAAUCACAUAUGCGAUGAUUUAAGCCAAUGCAUAAUUGAUUGUAAUGCUUGGCAAGCCGAUUUCGGUUUGCAUAAGCACAAGGCCAAGCAC--ACAGCAUCCAGUCUUCGGAAAUU ((((.(((((..((((((.((........)).))))))..)))))....((((((((((((((....))))))((....)).....)))))))).--...))))....((....)).... ( -35.20) >DroEre_CAF1 37246 120 - 1 AUGUAAAUCACGUAUGCGAUGAUUUAAGCCAAUGCAUAAUUGAUUGUAAUGCUUGGCAAGCCGAUUUCGGUUUGCAUAAGCACAAGGCCAAGCACACACAGCAUCCAGUCUUCGGAAAUU ...(((((((((....)).))))))).(((..((((........)))).((((((((((((((....)))))))).))))))...)))...((.......))......((....)).... ( -36.10) >DroYak_CAF1 35052 118 - 1 AUGUAAAUCACGUAUGCGAUGAUUUAAGCCAAUGCAUAAUUGAUUGUAAUGCUUGGCAAGCCGAUUUCGGUUUGCAUAAGCACAAGGCCAAGCAC--ACAGCAUCCAGUCUUCGGAAAUU ...(((((((((....)).))))))).(((..((((........)))).((((((((((((((....)))))))).))))))...)))...((..--...))......((....)).... ( -36.40) >consensus AUGUAAAUCACAUAUGCGAUGAUUUAAGCCAAUGCAUAAUUGAUUGUAAUGCUUGGCAAGCCGAUUUCGGUUUGCAUAAGCACAAGGCCAAGCAC__ACAGCAUCCAGUCUUCGGAAAUU .(((.(((((..((((((.((........)).))))))..)))))((..((((((((((((((....))))))((....)).....))))))))...)).)))(((.......))).... (-35.58 = -35.58 + -0.00)


| Location | 13,799,551 – 13,799,671 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.92 |
| Mean single sequence MFE | -22.32 |
| Consensus MFE | -20.00 |
| Energy contribution | -20.40 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.06 |
| SVM RNA-class probability | 0.998315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13799551 120 + 23771897 CUUAUGCAAACCGAAAUCGGCUUGCCAAGCAUUACAAUCAAUUAUGCAUUGGCUUAAAUCAUCGCAUAUGUGAUUUACAUGCACACACACACACACACACCCAAGGCAGUCAGUCGUAGC .....((((.(((....))).))))...((((...(((((..(((((..(((......)))..)))))..)))))...))))......................(((.....)))..... ( -24.40) >DroSec_CAF1 36393 108 + 1 CUUAUGCAAACCGAAAUCGGCUUGCCAAGCAUUACAAUCAAUUAUGCAUUGGCUUAAAUCAUCGCAUAUGUGAUUUACAUGCACACA--------CACACCCAAGGCAGU----CAUAGC (((..((((.(((....))).))))...((((...(((((..(((((..(((......)))..)))))..)))))...)))).....--------.......))).....----...... ( -22.80) >DroSim_CAF1 32579 108 + 1 CUUAUGCAAACCGAAAUCGGCUUGCCAAGCAUUACAAUCAAUUAUGCAUUGGCUUAAAUCAUCGCAUAUGUGAUUUACAUGCACACA--------CACACCCAAGGCAGU----CCUAGC .....((((.(((....))).))))...((((...(((((..(((((..(((......)))..)))))..)))))...)))).....--------........(((....----)))... ( -23.40) >DroEre_CAF1 37286 95 + 1 CUUAUGCAAACCGAAAUCGGCUUGCCAAGCAUUACAAUCAAUUAUGCAUUGGCUUAAAUCAUCGCAUACGUGAUUUACAUCCACACA--------CACACCCA----------------- ..(((((...(((....)))...(((((((((...........)))).)))))..........))))).(((.........)))...--------........----------------- ( -20.00) >DroYak_CAF1 35090 106 + 1 CUUAUGCAAACCGAAAUCGGCUUGCCAAGCAUUACAAUCAAUUAUGCAUUGGCUUAAAUCAUCGCAUACGUGAUUUACAUACACACA--------CACACAUA--GCACA----CAUAGC ....(((...(((....)))...(((((((((...........)))).))))).(((((((.((....)))))))))..........--------........--)))..----...... ( -21.00) >consensus CUUAUGCAAACCGAAAUCGGCUUGCCAAGCAUUACAAUCAAUUAUGCAUUGGCUUAAAUCAUCGCAUAUGUGAUUUACAUGCACACA________CACACCCAAGGCAGU____CAUAGC .....((((.(((....))).))))...((((...(((((..(((((..(((......)))..)))))..)))))...))))...................................... (-20.00 = -20.40 + 0.40)


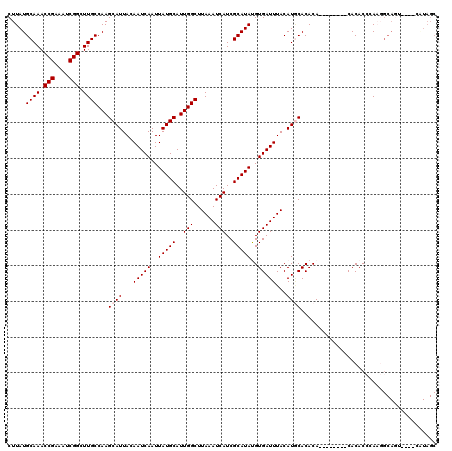
| Location | 13,799,551 – 13,799,671 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.92 |
| Mean single sequence MFE | -32.07 |
| Consensus MFE | -27.78 |
| Energy contribution | -27.86 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13799551 120 - 23771897 GCUACGACUGACUGCCUUGGGUGUGUGUGUGUGUGUGUGCAUGUAAAUCACAUAUGCGAUGAUUUAAGCCAAUGCAUAAUUGAUUGUAAUGCUUGGCAAGCCGAUUUCGGUUUGCAUAAG (((((((..((.(((.((((.(.......((((((((((.((....))))))))))))........).)))).)))...))..)))))..))...((((((((....))))))))..... ( -35.46) >DroSec_CAF1 36393 108 - 1 GCUAUG----ACUGCCUUGGGUGUG--------UGUGUGCAUGUAAAUCACAUAUGCGAUGAUUUAAGCCAAUGCAUAAUUGAUUGUAAUGCUUGGCAAGCCGAUUUCGGUUUGCAUAAG ((...(----((((...(.((((((--------((((((.((....)))))))))))..........(((((.((((.((.....)).)))))))))..))).)...))))).))..... ( -31.80) >DroSim_CAF1 32579 108 - 1 GCUAGG----ACUGCCUUGGGUGUG--------UGUGUGCAUGUAAAUCACAUAUGCGAUGAUUUAAGCCAAUGCAUAAUUGAUUGUAAUGCUUGGCAAGCCGAUUUCGGUUUGCAUAAG .(((((----.....))))).((((--------((((((.((....))))))))))))......(((((...((((........))))..)))))((((((((....))))))))..... ( -32.40) >DroEre_CAF1 37286 95 - 1 -----------------UGGGUGUG--------UGUGUGGAUGUAAAUCACGUAUGCGAUGAUUUAAGCCAAUGCAUAAUUGAUUGUAAUGCUUGGCAAGCCGAUUUCGGUUUGCAUAAG -----------------....((((--------(((.(((...(((((((((....)).)))))))..)))))))))).............((((((((((((....)))))))).)))) ( -30.60) >DroYak_CAF1 35090 106 - 1 GCUAUG----UGUGC--UAUGUGUG--------UGUGUGUAUGUAAAUCACGUAUGCGAUGAUUUAAGCCAAUGCAUAAUUGAUUGUAAUGCUUGGCAAGCCGAUUUCGGUUUGCAUAAG ...(((----..(((--.....)))--------..)))((((...(((((..((((((.((........)).))))))..)))))...))))...((((((((....))))))))..... ( -30.10) >consensus GCUAUG____ACUGCCUUGGGUGUG________UGUGUGCAUGUAAAUCACAUAUGCGAUGAUUUAAGCCAAUGCAUAAUUGAUUGUAAUGCUUGGCAAGCCGAUUUCGGUUUGCAUAAG ......................................((((...(((((..((((((.((........)).))))))..)))))...))))...((((((((....))))))))..... (-27.78 = -27.86 + 0.08)

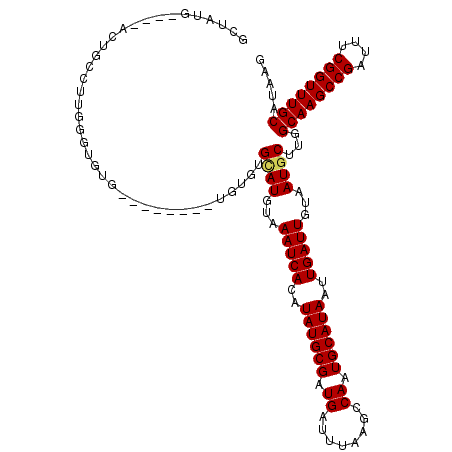

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:33 2006