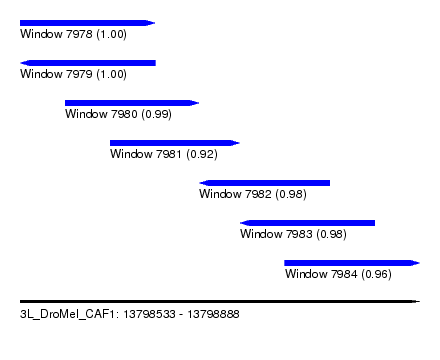
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,798,533 – 13,798,888 |
| Length | 355 |
| Max. P | 0.999803 |

| Location | 13,798,533 – 13,798,653 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -34.18 |
| Consensus MFE | -30.00 |
| Energy contribution | -30.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.88 |
| SVM decision value | 4.12 |
| SVM RNA-class probability | 0.999803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13798533 120 + 23771897 UCUUCCUCCUCCACCUGAUGUCCUUUUUCUAGGACUUUAGCUCUUGCAGAUUCCGCACACGUGCGUGUACCUUUGCUGCCAGGUUAAUGUCCUCAAACGGACAUGUCCUCCUAGGUCCCU ............(((((((((((.(((...(((((.(((((.((.((((.......((((....)))).......)))).))))))).))))).))).)))))).......))))).... ( -33.35) >DroSec_CAF1 35378 120 + 1 UCUUCUUCCUCCACCUGAUGUCCUUUUUCUAGGACUUUAGCUUUUGCAGAUUCCGCACACGUGCGUGUACCUUUGCUGCCAGGUUAAUGUCCUCAAACGGACAUGUCCACCCAGGUCCCU ............(((((((((((.(((...(((((.(((((((..((((.......((((....)))).......)))).))))))).))))).))).)))))).......))))).... ( -34.15) >DroSim_CAF1 31560 120 + 1 UCUUCCUCCUCCACCUGAUGUCCUUUUUCUAGGACUUUAGCUUUUGCAGAUUCCGCACACGUGCGUGUACCUUUGCUGCCAGGUUAAUGUCCUCAAACGGACAUGUCCACCCAGGUCCCU ............(((((((((((.(((...(((((.(((((((..((((.......((((....)))).......)))).))))))).))))).))).)))))).......))))).... ( -34.15) >DroEre_CAF1 36313 120 + 1 UCUUCCUACCCCACAUGAUGUCCUUUUUCUAGGACUUGGGCUCUUGCAGAUUCCGCACACGUGCGUGUACCUUUGCUGCCAGGUUAAUGUCCUCAAACGGACAUGUCCAACUAGGUCCCC ....((((...........(((((......)))))(((((((((.((((.......((((....)))).......)))).)))...((((((......)))))))))))).))))..... ( -34.84) >DroYak_CAF1 34161 120 + 1 UCUACCUCCUCCACAUGAUGUCCUUUUUCUAGGACCUUAGCUCUUGCAGAUUCCGCACACGUGCGUGUACCUUUGCUGCCAGGUUAAUGUCCUCCAACGGACAUGUCCACCUGGGUCCCC ...............(((.(((((......))))).)))......(((((..(((((....)))).)....)))))..((((((..((((((......))))))....))))))...... ( -34.40) >consensus UCUUCCUCCUCCACCUGAUGUCCUUUUUCUAGGACUUUAGCUCUUGCAGAUUCCGCACACGUGCGUGUACCUUUGCUGCCAGGUUAAUGUCCUCAAACGGACAUGUCCACCUAGGUCCCU ............(((((..(((((......)))))....((....(((((..(((((....)))).)....))))).))..((...((((((......))))))..))...))))).... (-30.00 = -30.00 + 0.00)


| Location | 13,798,533 – 13,798,653 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -38.38 |
| Consensus MFE | -33.86 |
| Energy contribution | -34.26 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.21 |
| SVM RNA-class probability | 0.998760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13798533 120 - 23771897 AGGGACCUAGGAGGACAUGUCCGUUUGAGGACAUUAACCUGGCAGCAAAGGUACACGCACGUGUGCGGAAUCUGCAAGAGCUAAAGUCCUAGAAAAAGGACAUCAGGUGGAGGAGGAAGA .....(((........((((((......))))))..(((((((((.....((((((....)))))).....))))..........(((((......)))))..)))))..)))....... ( -38.00) >DroSec_CAF1 35378 120 - 1 AGGGACCUGGGUGGACAUGUCCGUUUGAGGACAUUAACCUGGCAGCAAAGGUACACGCACGUGUGCGGAAUCUGCAAAAGCUAAAGUCCUAGAAAAAGGACAUCAGGUGGAGGAAGAAGA ....((((((((....((((((......))))))..)))..((((.....((((((....)))))).....))))..........(((((......)))))..)))))............ ( -38.20) >DroSim_CAF1 31560 120 - 1 AGGGACCUGGGUGGACAUGUCCGUUUGAGGACAUUAACCUGGCAGCAAAGGUACACGCACGUGUGCGGAAUCUGCAAAAGCUAAAGUCCUAGAAAAAGGACAUCAGGUGGAGGAGGAAGA ....((((((((....((((((......))))))..)))..((((.....((((((....)))))).....))))..........(((((......)))))..)))))............ ( -38.20) >DroEre_CAF1 36313 120 - 1 GGGGACCUAGUUGGACAUGUCCGUUUGAGGACAUUAACCUGGCAGCAAAGGUACACGCACGUGUGCGGAAUCUGCAAGAGCCCAAGUCCUAGAAAAAGGACAUCAUGUGGGGUAGGAAGA .....(((........((((((......))))))..((((.(((......((((((....))))))((..((.....))..))..(((((......)))))....))).))))))).... ( -37.60) >DroYak_CAF1 34161 120 - 1 GGGGACCCAGGUGGACAUGUCCGUUGGAGGACAUUAACCUGGCAGCAAAGGUACACGCACGUGUGCGGAAUCUGCAAGAGCUAAGGUCCUAGAAAAAGGACAUCAUGUGGAGGAGGUAGA (....)((((((....((((((......))))))..))))))........((((((....))))))....(((((.....(((..(((((......)))))......))).....))))) ( -39.90) >consensus AGGGACCUAGGUGGACAUGUCCGUUUGAGGACAUUAACCUGGCAGCAAAGGUACACGCACGUGUGCGGAAUCUGCAAGAGCUAAAGUCCUAGAAAAAGGACAUCAGGUGGAGGAGGAAGA .....((.........((((((......))))))..(((((((((.....((((((....)))))).....))))..........(((((......)))))..))))))).......... (-33.86 = -34.26 + 0.40)

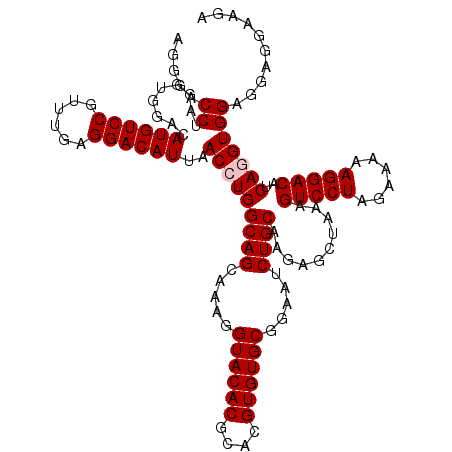
| Location | 13,798,573 – 13,798,692 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.62 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -33.30 |
| Energy contribution | -33.58 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13798573 119 + 23771897 CUCUUGCAGAUUCCGCACACGUGCGUGUACCUUUGCUGCCAGGUUAAUGUCCUCAAACGGACAUGUCCUCCUAGGUCCCUGGGGUAGCCAA-ACCAGUUAGCCAGCUGCCUCACAUUCUU .....((((.....((((((....))))......(((((((((...((((((......))))))..))).((((....))))))))))...-........))...))))........... ( -35.80) >DroSec_CAF1 35418 119 + 1 CUUUUGCAGAUUCCGCACACGUGCGUGUACCUUUGCUGCCAGGUUAAUGUCCUCAAACGGACAUGUCCACCCAGGUCCCUAGGGUAGCCAA-ACCAGUUAGCCAGCUGCCUCACAUUCUU .....((((....((((....)))).........((((.(.((((.((((((......))))))....((((((....)).)))).....)-))).).))))...))))........... ( -31.80) >DroSim_CAF1 31600 119 + 1 CUUUUGCAGAUUCCGCACACGUGCGUGUACCUUUGCUGCCAGGUUAAUGUCCUCAAACGGACAUGUCCACCCAGGUCCCUAGGGUAGCCAA-ACCAGUUAGCCAGCUGCCUCACAUUCUU .....((((....((((....)))).........((((.(.((((.((((((......))))))....((((((....)).)))).....)-))).).))))...))))........... ( -31.80) >DroEre_CAF1 36353 120 + 1 CUCUUGCAGAUUCCGCACACGUGCGUGUACCUUUGCUGCCAGGUUAAUGUCCUCAAACGGACAUGUCCAACUAGGUCCCCAGGGUAGCCAGCACCCGUUAGCCAGCUGCCUCACAUUCUU .....(((((..(((((....)))).)....))))).(((.((((.((((((......))))))....)))).))).....(((((((..((........))..)))))))......... ( -34.60) >DroYak_CAF1 34201 106 + 1 CUCUUGCAGAUUCCGCACACGUGCGUGUACCUUUGCUGCCAGGUUAAUGUCCUCCAACGGACAUGUCCACCUGGGUCCCCAGGGUAGC--------------CAGCUGCCUCACAUUCUU .....((((....((((....)))).........((((((.((...((((((......))))))..))..((((....))))))))))--------------...))))........... ( -37.50) >consensus CUCUUGCAGAUUCCGCACACGUGCGUGUACCUUUGCUGCCAGGUUAAUGUCCUCAAACGGACAUGUCCACCUAGGUCCCUAGGGUAGCCAA_ACCAGUUAGCCAGCUGCCUCACAUUCUU .....((((....((((....)))).........((((((.((...((((((......))))))..))..((((....)))))))))).................))))........... (-33.30 = -33.58 + 0.28)

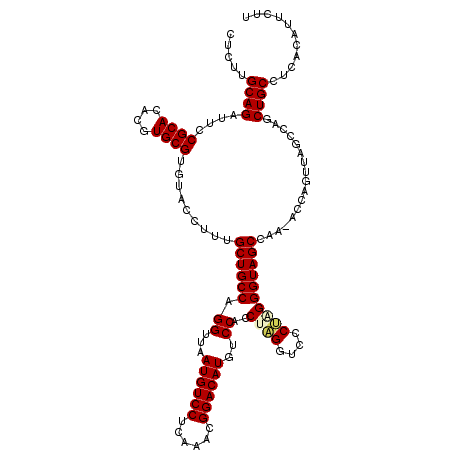
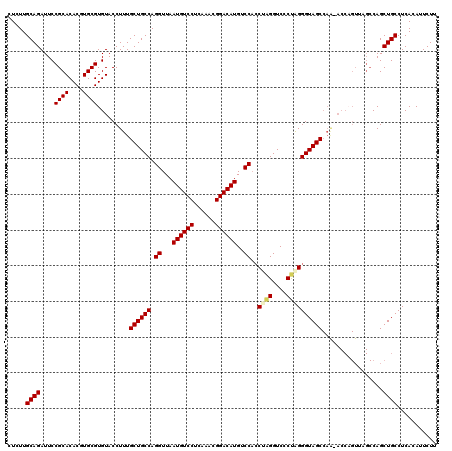
| Location | 13,798,613 – 13,798,728 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.59 |
| Mean single sequence MFE | -37.29 |
| Consensus MFE | -28.23 |
| Energy contribution | -28.15 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13798613 115 + 23771897 AGGUUAAUGUCCUCAAACGGACAUGUCCUCCUAGGUCCCUGGGGUAGCCAA-ACCAGUUAGCCAGCUGCCUCACAUUCUUUUUUUUUUUUAAUUGAGAGAGAUAGGAGACGGC----CGA .((((.((((((......))))))(((.(((((......(((((((((...-............)))))))))..(((((((..((....))..))))))).)))))))))))----).. ( -37.76) >DroSec_CAF1 35458 111 + 1 AGGUUAAUGUCCUCAAACGGACAUGUCCACCCAGGUCCCUAGGGUAGCCAA-ACCAGUUAGCCAGCUGCCUCACAUUCUUUU----UUUUAAUUGAGAGAGAUAGGAGACGGC----CGG .(((((((((((......)))).((.(.((((((....)).)))).).)).-....))))))).((((.(((....((((((----((......))))))))...))).))))----... ( -36.50) >DroSim_CAF1 31640 113 + 1 AGGUUAAUGUCCUCAAACGGACAUGUCCACCCAGGUCCCUAGGGUAGCCAA-ACCAGUUAGCCAGCUGCCUCACAUUCUUUUU--UUUUUAAUUGAGAGAGAUAGGAGACGGC----CGG .(((((((((((......)))).((.(.((((((....)).)))).).)).-....))))))).((((.(((....(((((((--(........))))))))...))).))))----... ( -36.30) >DroEre_CAF1 36393 111 + 1 AGGUUAAUGUCCUCAAACGGACAUGUCCAACUAGGUCCCCAGGGUAGCCAGCACCCGUUAGCCAGCUGCCUCACAUUCUUUU----UGUUAAUUGGGAGAGAUAGGAGACGGC----UG- .((((.((((((......))))))(((...(((..(((((((((((((..((........))..))))))).(((.......----)))....)))).))..)))..))))))----).- ( -35.20) >DroYak_CAF1 34241 102 + 1 AGGUUAAUGUCCUCCAACGGACAUGUCCACCUGGGUCCCCAGGGUAGC--------------CAGCUGCCUCACAUUCUUUU----UGUUAAUUGAGAGAGAUAGGAGAUAGGAGGCAGG .(((((((((((......)))))).....(((((....))))).))))--------------)..(((((((....((((((----......((....))...))))))...))))))). ( -40.70) >consensus AGGUUAAUGUCCUCAAACGGACAUGUCCACCUAGGUCCCUAGGGUAGCCAA_ACCAGUUAGCCAGCUGCCUCACAUUCUUUU____UUUUAAUUGAGAGAGAUAGGAGACGGC____CGG .((...((((((......))))))..))......((((((((((((((................)))))))....(((((((............))))))).)))).))).......... (-28.23 = -28.15 + -0.08)

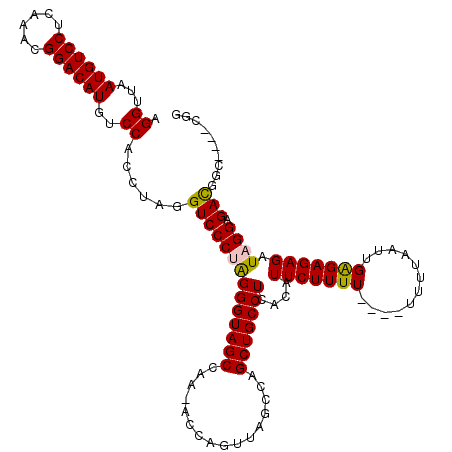
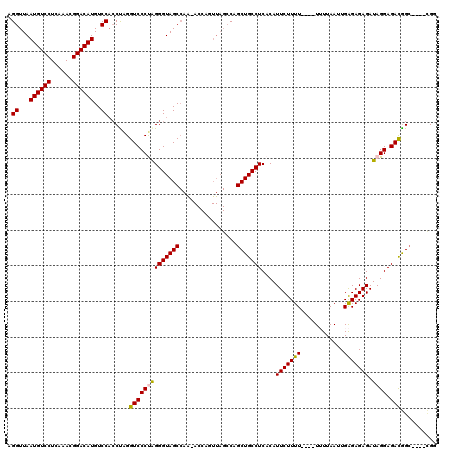
| Location | 13,798,692 – 13,798,808 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.75 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -25.95 |
| Energy contribution | -26.23 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13798692 116 - 23771897 ACAAUUGCAGGAUGCAGUUGUAGGCAGGAUGCUGCAGAUUCGGAAGUGUAAUUAUCCCCAUAUUCCAAUCUGGUGCUCCUUCG----GCCGUCUCCUAUCUCUCUCAAUUAAAAAAAAAA ((((((((.....)))))))).(((((((.((..((((((.(((((((..........))).))))))))))..))))))...----))).............................. ( -30.60) >DroSec_CAF1 35537 112 - 1 ACAAUUGCAGGAUGCAGUUGUAGGCAGGAUGCUGCAGAUUCGGAAGUGUAAUUAUCCCCAUAUUCCAAUCUGGUGCUCCUCCG----GCCGUCUCCUAUCUCUCUCAAUUAAAA----AA ((((((((.....)))))))).(((.(((.((..((((((.(((((((..........))).))))))))))..))...))).----)))........................----.. ( -30.80) >DroSim_CAF1 31719 114 - 1 ACAAUUGCAGGAUGCAGUUGUAGGCAGGAUGCUGCAGAUUCGGAAGUGUAAUUAUCCCCAUAUUCCAAUCUGGUGCUCCUCCG----GCCGUCUCCUAUCUCUCUCAAUUAAAAA--AAA ((((((((.....)))))))).(((.(((.((..((((((.(((((((..........))).))))))))))..))...))).----))).........................--... ( -30.80) >DroEre_CAF1 36473 111 - 1 ACAAUUGCAGGAUGCAGUUGUAGGCAGGAUGCUGCAGAUUCGGAAGUGUAAUUAUCCCCAUAUUCCAAUCUGGUGCUCCC-CA----GCCGUCUCCUAUCUCUCCCAAUUAACA----AA ((((((((.....)))))))).(((.(((.((..((((((.(((((((..........))).))))))))))..))))).-..----)))........................----.. ( -29.50) >DroYak_CAF1 34307 116 - 1 ACAAUUGCAGGAUGCAGUUGUAGGCAGGAUGCUGCAGAUUCGGAAGUGUAAUUAUCCCUAUAUUCCAAUCUGGUGCGCCUCCUGCCUCCUAUCUCCUAUCUCUCUCAAUUAACA----AA ((((((((.....))))))))(((((((((((..((((((.((((.((((........))))))))))))))..)))..))))))))...........................----.. ( -36.80) >consensus ACAAUUGCAGGAUGCAGUUGUAGGCAGGAUGCUGCAGAUUCGGAAGUGUAAUUAUCCCCAUAUUCCAAUCUGGUGCUCCUCCG____GCCGUCUCCUAUCUCUCUCAAUUAAAA____AA ((((((((.....)))))))).(((((((.((..((((((.(((((((..........))).))))))))))..)))))).......))).............................. (-25.95 = -26.23 + 0.28)

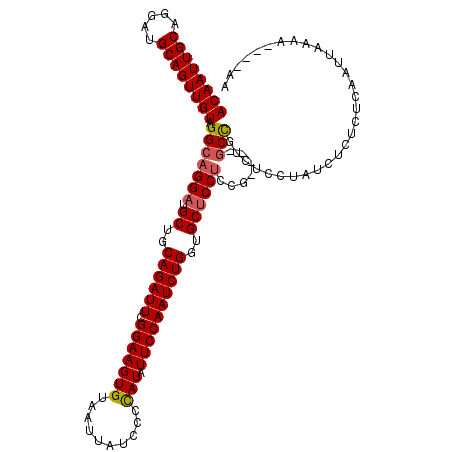
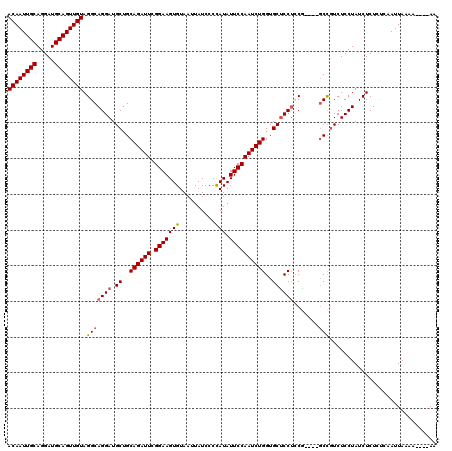
| Location | 13,798,728 – 13,798,848 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -32.52 |
| Consensus MFE | -30.72 |
| Energy contribution | -30.76 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13798728 120 - 23771897 UUCUCAAUUAAUUUUAAUUUCAUGUCAACACCUUCGGCCGACAAUUGCAGGAUGCAGUUGUAGGCAGGAUGCUGCAGAUUCGGAAGUGUAAUUAUCCCCAUAUUCCAAUCUGGUGCUCCU ....................................(((.((((((((.....)))))))).)))((((.((..((((((.(((((((..........))).))))))))))..)))))) ( -32.60) >DroSec_CAF1 35569 120 - 1 UUCUCAAUUAAUUUUAAUUUCAUGUCAACACCUUCGGCCGACAAUUGCAGGAUGCAGUUGUAGGCAGGAUGCUGCAGAUUCGGAAGUGUAAUUAUCCCCAUAUUCCAAUCUGGUGCUCCU ....................................(((.((((((((.....)))))))).)))((((.((..((((((.(((((((..........))).))))))))))..)))))) ( -32.60) >DroSim_CAF1 31753 120 - 1 UUCUCAAUUAAUUUUAAUUUCAUGUCAACACCUUCGGCCGACAAUUGCAGGAUGCAGUUGUAGGCAGGAUGCUGCAGAUUCGGAAGUGUAAUUAUCCCCAUAUUCCAAUCUGGUGCUCCU ....................................(((.((((((((.....)))))))).)))((((.((..((((((.(((((((..........))).))))))))))..)))))) ( -32.60) >DroEre_CAF1 36504 120 - 1 UUCUCAAUUAAUUUUAAUUUCAUGUCAACACCUUCGGCCGACAAUUGCAGGAUGCAGUUGUAGGCAGGAUGCUGCAGAUUCGGAAGUGUAAUUAUCCCCAUAUUCCAAUCUGGUGCUCCC ....................................(((.((((((((.....)))))))).))).(((.((..((((((.(((((((..........))).))))))))))..))))). ( -31.70) >DroYak_CAF1 34343 120 - 1 UUCUCAAUUAAUUUUAAUUUCAUGUCAACACCUUCGGCCGACAAUUGCAGGAUGCAGUUGUAGGCAGGAUGCUGCAGAUUCGGAAGUGUAAUUAUCCCUAUAUUCCAAUCUGGUGCGCCU ....................................(((.((((((((.....)))))))).)))(((.(((..((((((.((((.((((........))))))))))))))..)))))) ( -33.10) >consensus UUCUCAAUUAAUUUUAAUUUCAUGUCAACACCUUCGGCCGACAAUUGCAGGAUGCAGUUGUAGGCAGGAUGCUGCAGAUUCGGAAGUGUAAUUAUCCCCAUAUUCCAAUCUGGUGCUCCU ....................................(((.((((((((.....)))))))).))).(((.((..((((((.(((((((..........))).))))))))))..))))). (-30.72 = -30.76 + 0.04)

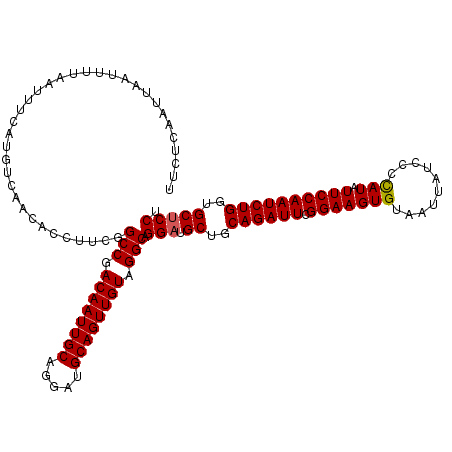
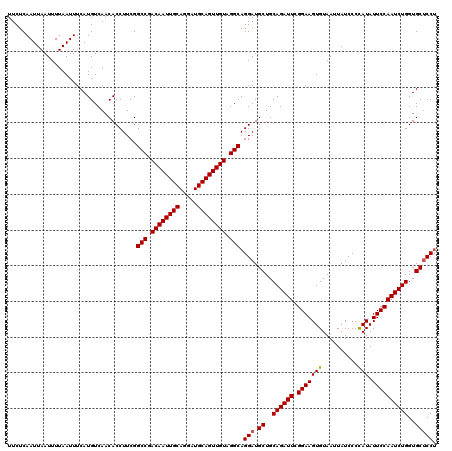
| Location | 13,798,768 – 13,798,888 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.33 |
| Mean single sequence MFE | -31.16 |
| Consensus MFE | -30.40 |
| Energy contribution | -30.60 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13798768 120 + 23771897 AAUCUGCAGCAUCCUGCCUACAACUGCAUCCUGCAAUUGUCGGCCGAAGGUGUUGACAUGAAAUUAAAAUUAAUUGAGAAUUCCGUGCACAUAGCAACGCAAUCGUGUCGACCAGCAACU ....(((.....((((((.((((.(((.....))).)))).)))...))).(((((((((((((((....)))))........(((((.....)).)))...))))))))))..)))... ( -32.30) >DroSec_CAF1 35609 120 + 1 AAUCUGCAGCAUCCUGCCUACAACUGCAUCCUGCAAUUGUCGGCCGAAGGUGUUGACAUGAAAUUAAAAUUAAUUGAGAAUUCCGUACACAUAGCAACGCAAUCGUGUCGUCCAGCAACU ....(((((((.((((((.((((.(((.....))).)))).)))...)))))))((((((((((((....)))))..................((...))..))))))).....)))... ( -28.40) >DroSim_CAF1 31793 120 + 1 AAUCUGCAGCAUCCUGCCUACAACUGCAUCCUGCAAUUGUCGGCCGAAGGUGUUGACAUGAAAUUAAAAUUAAUUGAGAAUUCCGUGCACAUAGCAACGCAAUCGUGUCGUCCAGCAACU ....(((((((.((((((.((((.(((.....))).)))).)))...)))))))((((((((((((....)))))........(((((.....)).)))...))))))).....)))... ( -31.70) >DroEre_CAF1 36544 120 + 1 AAUCUGCAGCAUCCUGCCUACAACUGCAUCCUGCAAUUGUCGGCCGAAGGUGUUGACAUGAAAUUAAAAUUAAUUGAGAAUUCCGUGCACAUAGCAACGCAAUCGUGUCGUCCAGCAACU ....(((((((.((((((.((((.(((.....))).)))).)))...)))))))((((((((((((....)))))........(((((.....)).)))...))))))).....)))... ( -31.70) >DroYak_CAF1 34383 120 + 1 AAUCUGCAGCAUCCUGCCUACAACUGCAUCCUGCAAUUGUCGGCCGAAGGUGUUGACAUGAAAUUAAAAUUAAUUGAGAAUUCCGUGCACAUAGCAACGCAAUCGUGUCGUCCAGCAACU ....(((((((.((((((.((((.(((.....))).)))).)))...)))))))((((((((((((....)))))........(((((.....)).)))...))))))).....)))... ( -31.70) >consensus AAUCUGCAGCAUCCUGCCUACAACUGCAUCCUGCAAUUGUCGGCCGAAGGUGUUGACAUGAAAUUAAAAUUAAUUGAGAAUUCCGUGCACAUAGCAACGCAAUCGUGUCGUCCAGCAACU ....(((((((.((((((.((((.(((.....))).)))).)))...)))))))((((((((((((....)))))........(((((.....)).)))...))))))).....)))... (-30.40 = -30.60 + 0.20)

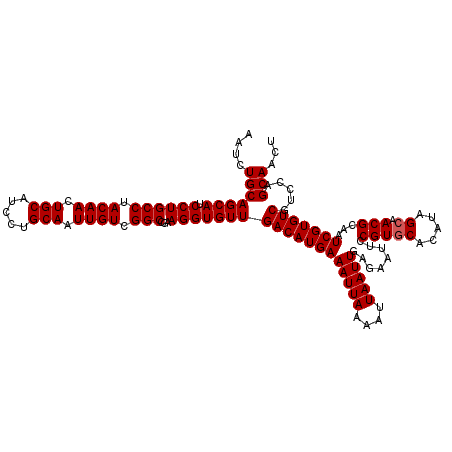
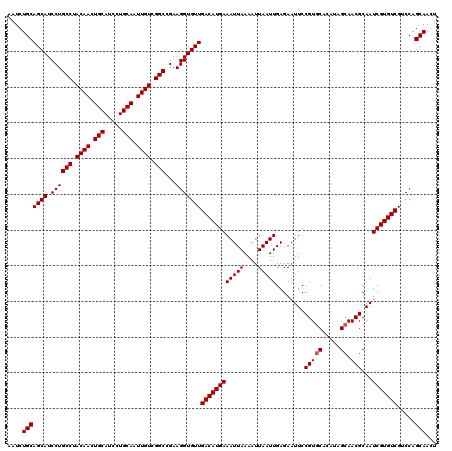
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:25 2006